![Scientists have discovered that two-thirds of the malaria parasite's genes are essential for normal growth, meaning there are many more possible drug targets than previously thought. [NIH]](https://genengnews.com/wp-content/uploads/2018/08/20130819malaria1099415684-1.jpg)
Scientists have discovered that two-thirds of the malaria parasite’s genes are essential for normal growth, meaning there are many more possible drug targets than previously thought. [NIH]
The creation of the aerosol can, the transfer of the Panama Canal construction to the U.S., and the formation of the CDC are all a direct result of malaria’s impact on human history, which cannot be overstated. Moreover, this single-cell parasite has been estimated to have caused the death of almost half of the world’s population since the Stone Age and has directly influenced human evolution through natural selection from diseases such as sickle cell. While this scourge on humanity has fostered some keen scientific minds and innovations, in recent years the threat of rising drug resistance has caused many malaria researchers to re-evaluate current strategies to combat his deadly disease.
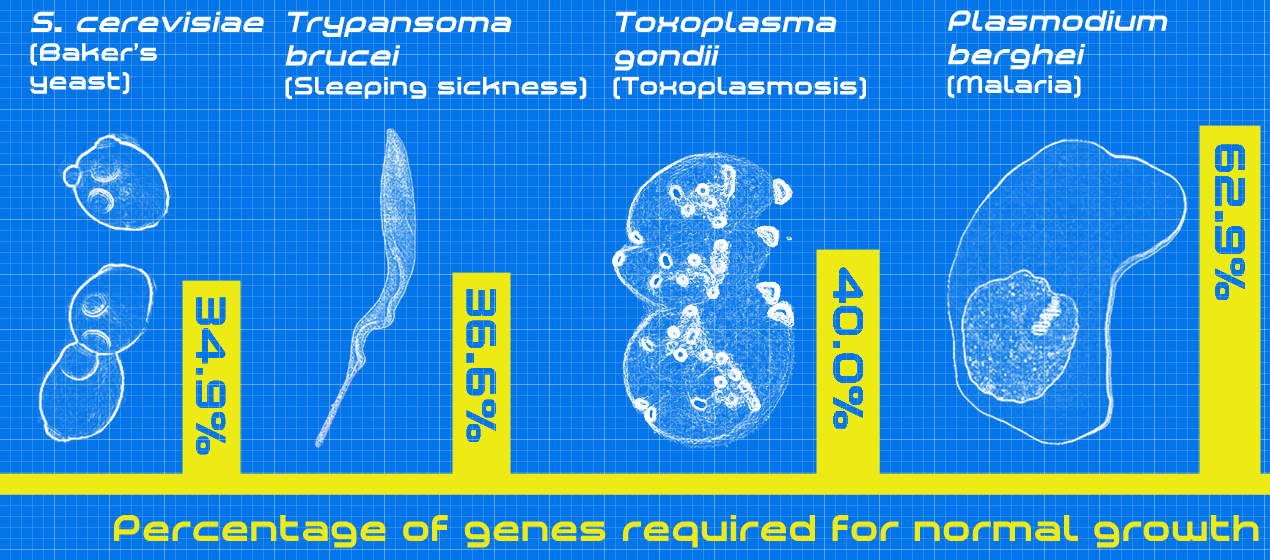
To that end, a team of investigators led by scientists at the Wellcome Trust Sanger Institute has just published one of the largest studies on malaria gene function, analyzing more than half of the genes in the parasite's genome. Findings from the new study—published recently in Cell in an article entitled “Functional Profiling of a Plasmodium Genome Reveals an Abundance of Essential Genes”—showed that two-thirds of these genes the researchers analyzed were essential for survival—the largest proportion of essential genes found in any organism studied to date. These results could aid in the identification of many potential targets for new antimalarial drug development.
“This study of unprecedented scale has resulted in many more unique drug targets for malaria,” remarked Francisco Javier Gamo, Ph.D., director of the malaria unit at GlaxoSmithKline and not directly involved in the current study. “The Holy Grail would be to discover genes that are essential across all of the parasite lifecycle stages, and if we could target those with drugs it would leave malaria with nowhere to hide. The technology that the Sanger Institute has developed gives us the potential to ask those questions systematically for the first time.”
Since roughly half of the genes in the Plasmodium genome have no homologs in another organism, deciphering the parasite’s genetics has been particularly challenging. This new study, however, provides the first ever experimental evidence of function for most of those genes.
Using a mouse model of malaria infection, Plasmodium berghei, the investigators designed a new method to decipher the function of the malaria parasite's genes. The team switched off, or knocked out, 2578 genes—more than half of the genome—and gave each knockout a unique DNA barcode. The barcoding method involves tagging specific genes with a molecular ID, enabling scientists to identify individual mutants and measure the growth of the genetically modified parasite by counting the number of barcodes using a benchtop sequencer.
Source: Wellcome Sanger Institute
The researchers used next-generation sequencing technology to count those barcodes, and hence measure the growth of each genetically modified malaria parasite. If the switched-off gene was not essential, the parasite numbers shot up, but if the knocked-out gene was essential, the parasite disappeared.
“This work was made possible by a new method that enabled us to investigate more than 2500 genes in a single study—more than the entire research community has studied over the past two decades,” explained co-senior study investigator Oliver Billker, Ph.D., senior group leader at Sanger. “We believe that this method can be used to build a deep understanding of many unknown aspects of malaria biology and radically speed up our understanding of gene function and prioritization of drug targets.”
The researchers systematically showed that the malaria parasite can easily dispose of the genes that produce proteins that give away its presence to the host's immune system. This poses problems for the development of malaria vaccines, as the parasite can quickly alter its appearance to the human immune system, and, as a result, the parasite can build resistance to the vaccine.
“We knew from previous work that on its surface the malaria parasite has many dispensable parts,” remarked co-senior study investigator Julian Rayner, Ph.D., senior group leader and director of Wellcome Genome Campus Connecting Science. “Our study found that below the surface the parasite is more of a Formula 1 race car than a clunky people carrier. The parasite is fine-tuned and retains the absolute essential genes needed for growth. This is both good and bad: The bad news is it can easily get rid of the genes behind the targets we are trying to design vaccines for, but the flip side is there are many more essential gene targets for new drugs than we previously thought.”
The authors stressed that the factors influencing gene function go well beyond the realm of basic science research, concluding that “the level of genetic redundancy in a single-celled organism may thus reflect the degree of environmental variation it experiences. In the case of Plasmodium parasites, this helps rationalize both the relative successes of drugs and the greater difficulty of making an effective vaccine.”