February 1, 2013 (Vol. 33, No. 3)
David Daniels, Ph.D.
Biomarker research and development has evolved over the past years from looking for a single marker (e.g., PSA) linked to a disease state to looking for a panel of markers that can capture the heterogeneity inherent in both the disease and the impacted patient population.
That is one of the key messages to be delivered at GTC’s “Biomarkers Summit” next month. Across the board, resources are being focused on the delivery of more precise, quantifiable biomarkers with predictive value in therapeutic decisions and for the prognosis of illness.
“Our focus on biomarker development is the recognition that the new products need to provide cost savings for the already strapped healthcare systems rather than just be cost effective,” shares Paul Billings, M.D., Ph.D., CMO at Life Technologies.
“We have built a new medical sciences group to address the needs of the multiple delivery systems in the world—from the sophisticated medical clinics in the developed world to the nurse-run shanty clinic in the third world. Providing tools for equitable access to quality diagnosis, on assay platforms that can provide care for all patients, is our goal.”
Life Tech’s medical sciences division has been built by acquisition of Pinpoint Genomics, Navigenics, and Compendia, and collaborations with partners such as Ingenuity Systems and CollabRx. The division is focused on taking the tools that have been used in the life science laboratories and providing molecular diagnostic data to the clinic. The intent is to deliver data in a valuable format that can be used by the molecular pathologist or the treating physician.
The division is developing the Pervenio™ Lung RS assay, a 14-gene expression profile that serves as a risk stratifier that uses a weighted algorithm for the expressed biomarkers within the tumor biopsy, a first-of-its-kind prognostic test for lung cancer, the firm reports.
Initially, tests will be offered as a service through Life Tech’s CLIA laboratory. Then, from the performance lessons learned, Life Tech’s will develop a simpler assay platform, with FDA approval, that can be dispersed globally without reduction of the essential content in the biomarker panel. The focus is on the workflow—screening for known mutations using established easy-to-use assay platforms, like RT-PCR. Should the screen not produce useful results, clinicians can search for new mutations via discovery platforms like next-gen sequencing (NGS).
At Sequenom, the company provides both the tools (DNA mass spectrometry and reagents) for confirmatory biomarker development as well as serving on the front lines as a diagnostic service provider (CLIA lab). The beauty of DNA mass spec is that it can process multiplexed PCR samples (10–60 loci) in a method that is quantitative when used for profiling tumor biopsies that are either archival or fresh tissue.
Given a tumor sample with multiple somatic mutations, the instrument enables the determination of the homogeneity of the cells, in which case the mutations will have the same allele frequency. Accuracy, as measured by coefficient of variance, is less than 2%. Despite this level of sensitivity, the mass spec can only be used as a confirmatory tool looking for known mutations. Discovery is best done using DNA sequencing. DNA mass spec can also be used to study methylation in tumor samples.
“In the not-too-distant future, we will be looking for mutations in plasma samples rather than biopsies,” predicts Charles Cantor, Ph.D., CSO at Sequenom.
“The key is to look noninvasively for mutations within plasma samples such that we can potentially catch the disease state earlier, rather than after tumor formation. Regardless of the tumor type, this approach will enable us to monitor therapeutic response and metastatic potential noninvasively. DNA mass spec is an ultrasensitive detection product that can detect somatic mutations at levels of 1 per 1,000. This level of sensitivity is critical for the future of plasma screening. NGS technology is not that sensitive.”
Sequenom’s CLIA lab is using automated DNA mass spec to provide three different test protocols: (1) carrier screening for cystic fibrosis looking at more than 100 different mutations, (2) adult macular degeneration progression using an SNP test with 13 loci, and (3) a noninvasive test for Rh compatibility between a mother and her unborn fetus.
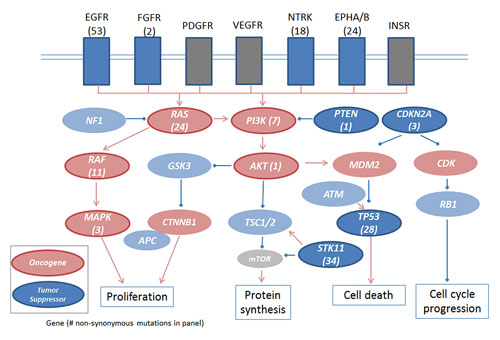
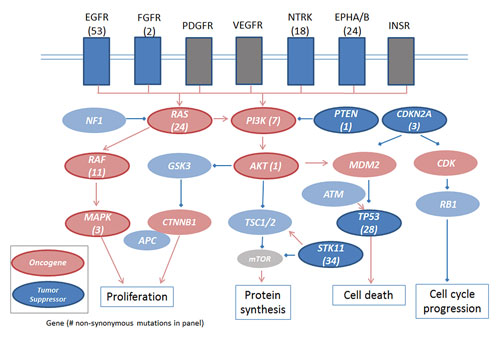
Sequenom’s LungCarta panel of 214 somatic mutations in 26 tumor suppressors and oncogenes covers highly mutated pathways in lung adenocarcinomas.
Sequenom has also set up an NGS facility within a CLIA lab in San Diego using Illumina’s HiSEQ platform. The NGS platform has been set up for noninvasive aneuploidy detection of maternal plasma (10 cc sample) looking at chromosomes 13, 18, and 21. The lab says it has analyzed more than 40,000 samples this year and is planning to increase that volume up to 100,000 samples per year. Most of these samples come from the U.S., but given the development of a new blood collection tube that allows for 72-hour ambient shipping, the lab is looking to increase the number of samples from outside the U.S.

Scientists are using Illumina’s HiSeq system to discover molecular biomarkers that may provide opportunities for early detection of a range of diseases.
Drug Development
During drug development, biomarkers function as pharmacodynamic markers to help assess the mechanism of action of a drug candidate, to define the downstream biological pathway, and to determine whether the drug is engaging the target with the anticipated biological effect. Later, biomarkers help determine whether a drug is effective using the tested regime (route of delivery, dosage level, and length of exposure time).
Following early development, the second stage is to use biomarkers to help segment patients for clinical trials. Part of the consideration here is how heterogeneous the disease is; are there homogeneous subsets of patients that will respond differentially to the drug based on different mechanisms of the disease?
“Biomarker research is focused on on- target effects,” says Nick Dracopoli, Ph.D., vp, head of oncology biomarkers at Janssen Research and Development, a J&J company.
“We look at indications and at patients with those indications that are most likely to respond to the drug candidates we’re developing. For oncology biomarkers, germ-line effects are weaker indicators than somatic changes in the tumor. As a consequence, SNP-based, genome-wide association studies are not very useful. It is better to focus on molecular changes within the tumor and define gene expression profiles and epigenetic modifications that correlate with the tumor phenotype. We are increasingly tracking patient immune response, particularly as more immuno-oncology products are moving into the drug development pipeline.”
The number of biomarkers being developed varies from project to project. But it is very clear that to be successful in the clinic, the biomarkers and the assays need to be of low complexity. Of the 10 to 12 companion diagnostics that have been approved by the FDA to date, all measure the status of the drug target (on-target markers). For example, EGFR measures the level of receptor expression; Braf and Kras markers measure the presence of the mutation and translocation in the ALK gene measures gene knockout.
It is important to realize that molecular profiles for first-in-class drugs are not optimal because they are based on only a few patients. Consequently they have weak predictive value overall.
“Aside from that rule of thumb, if you have a greater than 50% response rate for your drug, it is unlikely that you need a biomarker to predict response. Biomarker utility is best for drugs that would have a difficult road to approval, where it is critical to enrich for the subpopulation of responders. For example, Pfizer’s crizontinib was approved for non-small-cell lung patients but is only effective for 5% of all patients. If Pfizer was unable to demonstrate the relationship between activation of the ALK gene and disease, this inhibitor would not have been approved,” says Dr. Dracopoli.
“Drugs that are more broadly active can come to market without a companion diagnostic test. There is always a balance between the predictive values of the biomarker test and the response rates to treatment. That is, we should not treat if the chance of response is only 3–5%, rather than if it were 50% where the patient would want to take the chance if the drug were safe.”
An important take-home message is that mutations are not unique to an indication. So if you find a driver mutation in indications for which the drug has not been approved, you could discover new indications for the drug.
“At the end of the day, this is what cancer is—heterogeneous,” says Dr. Dracopoli. “We’d all love to treat one cancer with one drug and at one dose, but the story is more complex. The future of oncology is around understanding the molecular heterogeneity or underlying molecular pathology of the disease and the diversity of it, and then treat each patient accordingly.”
Clinical Considerations
“Given the complexity of biology,” says Achim Plum, Ph.D., principal consultant, Siemens, “whether is it cancer, metabolic disease, or any other disease state, we have been forced to move away from the idea that a single biomarker can capture the entire ‘story’ or mechanistic view of any disease. Hence newly developed biomarkers will be made up of a panel of markers that serve as a profile. In addition, with the sheer volume of DNA and protein analytics data, the clinic will need to employ software tools and algorithms to help the decision making.”
The task of getting broad profiling technologies that are analytical into a clinical setting and making them routine is difficult but not insurmontable. This will take a collaborative effort, something that Siemens among others are looking to develop. The key is to avoid technology hype and to establish good reliable software to process the data for decision making. “Data is not knowledge, and knowledge is not automatically decision making.”
As an academic, Daniel Chan, Ph.D., has a view of the whole value chain for biomarkers from discovery to development to use in the clinic. Dr. Chan holds the titles of professor in pathology, oncology, radiology, and urology, and is the director of the clinical chemistry division lab at Johns Hopkins Hospital.
Given his perspective from discovery to clinical use, Dr. Chan indicated that from the clinical point of view, “we need more markers.” He oversees the discovery of new biomarkers in his research lab, their validation in his translational research lab, and finally their utility in practice in his clinical chemistry lab. He is a strong advocate for collaboration of biomarker development from discovery to verification and validation to incorporation within the clinical practice.
Beyond the use of biomarkers for patient stratification and correlation between marker and therapeutic choice, as is the focus of the biopharma industry, for the clinic the use of biomarkers is for prevention and early detection. The earlier the detection, the better the outcome. That is, provide the “cure” before you need to initiate treatment.
To be successful in the future of biomarkers, we need to look beyond the biopharma focus and expand the horizon for early detection and monitor therapy later, says Dr. Chan. He describes a roadmap of developing bridges (to bridge the knowledge gaps), gates (decision gates for go/no go decisions as to whether a development path is viable), and partnerships (to collaborate with different points of view) for efficient new biomarker development.
According to Dr. Chan, we must define the intended use of the biomarker, which identifies the specific application and sets up the clinical study and study population to meet the clinical needs. We need to define specific assays to monitor biomarkers that will work within a clinical setting, not a research lab setting that uses disease models (tissue culture cells or small animals) and not real patient samples.
“The days when single markers are sufficient (PSA for prostate cancer or troponin for cardiovascular disease) are behind us. We need to develop a panel of markers or a profile pattern to address patient population heterogeneity and disease complexity that will guide our decision-making process,” remarks Dr. Chan. “Molecular biomarkers are giving way to protein biomarkers,” he adds.
Prevention and early detection will require the use of whole-body scans, so the sampling technology and analytical tools to be developed are critical to realize this goal. Assay ease of use, automation, and analytical performance that is suitable for the clinical lab are fundamental.
“An important future goal for biomarkers,” says Dr. Billings, “is to sample circulating tumor cells or circulating DNA in blood or plasma samples as a noninvasive measure of patient status. A decline in tumor biomarkers during chemotherapy, for example, could reflect the efficacy of the therapy. In contrast, an increase in tumor biomarkers, in a patient who had previously undergone surgery and therapy, might indicate disease recurrence, and is likely to do so before a tumor mass is detectable by imaging methods.”