January 15, 2013 (Vol. 33, No. 2)
Mitzi Perdue
Mathematics + 21st Century Computational Power = Safer and More Effective Healthcare
Eric Schadt, Ph.D., may be a scientist, and he may be the director of the $100 million enterprise, The Icahn Institute for Genomics and Multiscale Biology at Mount Sinai, but these days he’s thinking like a hedge fund manager.
However, even though he has $50 million worth of computational and data cyberinfrastructure available to him at Mount Sinai, he is definitely not acting like a hedge fund manager. That is, he’s not using the extraordinary computing power and mathematical techniques available at the institute to make real-time decisions on what stocks to buy, sell, or hold.
“Instead of asking what companies to bet on,” he explained in a recent interview, “we’re using these mathematical techniques to bet on which patients require treatment, and for those requiring treatment, what’s the best treatment for them.”
He expects that using the mathematical techniques developed for Wall Street, that in five to ten years, “Medicine will be so personalized, and the healthcare provider will have such a fine-grained understanding of what has perturbed the network of networks that resulted in the individual’s disease, that the physician will have both the knowledge to predict the course of the individual’s disease and the tools to treat or even prevent it.”
Ultimately, Dr. Schadt expects this mathematical approach to reduce humankind’s overall disease burden.

Dr. Eric Schadt is an expert on the generation and integration of large-scale sequence variation, molecular profiling, and clinical data in disease populations.
No Simple Linear Pathways in Biology
Why have the mathematical techniques used in finance suddenly become so important in medicine? According to Dr. Schadt, for most of mankind’s history, we’ve tried to explain the complex and the incomprehensible by telling ourselves fairly straight-forward linear stories, the kind that don’t require a lot of math. We explained lightning as Zeus hurling light bolts from the sky, or more recently, we explained type 2 diabetes as a linearly ordered pathway, starting with a defective gene and ending with the disease.
However, people with the same defective gene, whether it’s a gene for type 2 diabetes or virtually any other disease, often have remarkably different outcomes. With many single-gene disorders, there is a spectrum of phenotypic expressions, with some individuals barely touched by the disease while others experience the most extreme and horrific versions of it.
The linear explanation doesn’t account for this variety of outcomes, and it can’t because it doesn’t take into account the full, nonlinear complexity of diseases.
“DNA variation,” notes Dr. Schadt, “is just one dimension among many that define living systems. DNA doesn’t directly cause disease, but instead it has effects at the molecular level. It changes transcription. It changes proteins and it changes metabolite levels.
“Those different variables aren’t acting in isolation. They’re acting in a network, and it’s this network that senses the genetic and environmental perturbations that cause shifts in the system that lead to disease.”
He goes on to say that it requires the tools of advanced mathematics and unprecedented computing power to take the deeper snapshots of biological processes needed for constructing and generating models that will predict states of the system.
The phrase “multiscale biology” in the institute’s name comes from its goal of modeling the complexity of living systems, understanding not only all of the processes taking place at the molecular, cellular, tissue, organ, organismal, and community levels, but also how information flows between these levels.
“We generate big data (DNA sequencing, RNA sequencing, and so on), but then we also pull in lots of big data generated by others,” says Dr. Schadt. “We then have access to all of the clinical data from electronic medical records, and then we integrate the data to build complex network models that can tell us how best to diagnose and treat disease.”
Dr. Schadt points out that, for example, in the case of an individual with liver cancer, “The information on the liver tumor for just this one individual can lead to a terabyte or more of raw data. However, if we’re trying to find out where a given individual’s sequence is abnormal, we may want to compare it to similar information from 1,000 other people. Now you have a petabyte of information—that is one followed by 15 zeroes—that has to be analyzed for just one type of cancer.”
And for a still deeper understanding, researchers need to look at a hierarchy of levels, at multiple scales, from the small molecular level to the cellular to the tissue to the organism and then up to the large community level.
“As we move away from single genes and into networks, the complexity increases exponentially, and the scales of data we generate will exceed everything that is in the digital universe today,” continues Dr. Schadt. “We used to think a terabyte was a big number, and now we have petabytes and even exabytes of data.”
Disrupted Molecular Networks
The goal of Dr. Schadt and his colleagues is to tie all this data together to create more accurate models of diseases.
“We know that in the case of an individual, the disease is likely to have been caused by many changes in many genes that disrupted many different networks defining biological processes, and the goal is to discover and understand these disruptions.
“After we increase our understanding of the diseases,” he continues, “the question will be how to counter the perturbations in the disease-associated networks that operate in a network of networks. With this new information, derived from advanced computational techniques, we should be able to predict the current state of an individual’s disease, how likely it is to progress in the next year, what treatments are most likely to help his or her subtype, and which targets we can develop for new therapies that would be effective on the individual’s subtype.”
Dr. Schadt has an example of how, in the last four years, these new approaches have worked in practice. Our increased knowledge of the molecular causes of diabetes has changed how scientists and clinicians look at that disease. Previously, researches believed the major cause of diabetes was insulin resistance, and the treatment was to lower glucose levels.
“We now have found, by uncovering many of the molecular causes of diabetes, that most common forms of diabetes are related to beta cell dysfunction, and are more akin to type 2 diabetes,” he explains. “The pressing questions now are, how do we proliferate more normal beta cells and how do we prevent inflammatory processes from adversely affecting beta cell function and/or proliferation?”
With this and other knowledge, Dr. Schadt hopes that scientists will discover, on an individualized basis, that there are food types or diets that might correct the course of an individual’s type 2 diabetes. For instance, different types of sugar or fats have different effects on molecular functioning.
“The goal is to build maps of perturbagens that will cause changes in an individual’s molecular functioning, and with this, discover if the individual is on a healthy or unhealthy course, or if they already have the disease, what can we do to get the individual out of the disease state,” he says.
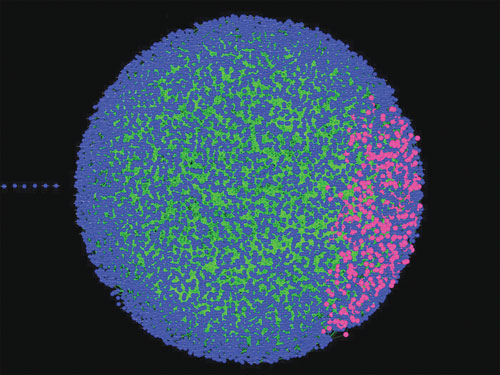
Omental Adipose Molecular Network: A causal gene network constructed from omental adipose tissue collected from individuals in a morbidly obese cohort (body mass index > 45). The blue/pink balls (or nodes) represent genes that were monitored in omental fat tissue across 800 individuals. Edges between the nodes represent causal associations between the corresponding genes. All of the edges in the network are directed, meaning they convey causal information regarding what genes cause others to change. The pink nodes in the network represent a special inflammatory/immune subnetwork of interest that we have identified in many studies as causal for many common human diseases such as Alzheimer’s disease, obesity, diabetes, heart disease, asthma, COPD, IBD, and different types of cancer.
Key Challenge
For Dr. Schadt, one of his biggest challenges is how to unify mathematically different modeling approaches to develop better models of disease. As he sees it, data-driven, hypothesis-free modeling approaches, i.e., structured learning approaches that assume we don’t know the rules of complex systems but must learn them from big data, are largely pursued independently of more hypothesis-driven modeling, i.e., approaches for which we assume we know the rules and how things are connected.
“We are seeking to integrate these approaches so that we get the best of both worlds while minimizing the weaknesses of each,” notes Dr. Schadt.
Dr. Schadt has some impressive resources available for his quest. The institute currently uses roughly 40,000 square feet, which includes a CLIA-certified sequencing core, a super computer (Minerva), wetlabs for sample preparation and running of molecular biology experiments, and dry lab space for different computational groups (statistical genetics, bioinformatics and sequence informatics, and network modeling/systems biology).
The institute also has some unique capabilities, including computing power that can manage petabytes [one quadrillion (1015) bytes] scales of data, and the presence of worldclass information technology personnel.
About half of the institute’s current faculty of 30 are experts in network modeling, predictive modeling, or machine learning. The other half is focused on sequence informatics, disease biology, and building interfaces. Dr. Schadt hopes to create the right ecosystem so that the diversity of talent across disciplines is all in the same space, learning and working with each other.
He’s still looking for additional talent. “We have staff scientist positions, faculty positions, post-doctoral positions, and we are recruiting students for our computational biology Ph.D. program.”
Interestingly, five years ago virtually none of these areas of expertise would have been required in any medical center. Today, they are essential to one of the institute’s key missions: handling large volumes of data, with the goal of developing information and creating understanding that can be translated rapidly into the clinical setting.
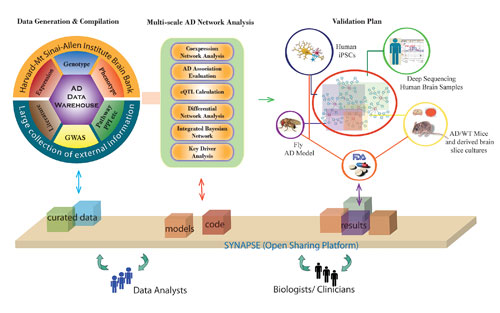
The workflow that the Icahn Institute for Genomics and Multiscale Biology employs to construct predictive models of disease and then to use those models to identify drug targets and biomarkers. In this example, the goal is to elucidate the complexity of Alzheimer’s.
Biobank Facility
Mount Sinai’s biobank is another resource available to the institute. Dr. Schadt is impressed that Irwin Bottinger, M.D., head of the biobank, made the far-sighted decision to change the biobank’s tissue collection efforts to an opt-in system in which, instead of de-identifying and aggregating the information, every tissue donor specifically gives his or her consent.
Since consent was required, it took longer to build the database to its current size of nearly 30,000 tissue donors, but the enormous benefit is that the institute’s researchers can go back to the tissue donors. Because the data is linked to the donor, it’s possible to find the ideal research candidates for a particular study.
In the case of those databanks that have chosen the opt-out model, they may have been able to acquire large numbers of tissue donors quickly, but by de-identifying the donors, the link between the individual and the information is broken, making the information far less useful.
The data Mount Sinai collects is also valuable because the population it serves is unusually diverse. Last year, the Mount Sinai Hospital treated nearly 60,000 inpatients and there were more than 500,000 outpatient hospital visits. The population the hospital draws from includes one of the country’s poorest zip codes on its northern border and, to the south, the population comes from one of the richest zip codes. The data collected reflects great socio-economic and ethnic diversity.
“When we want to understand different diseases, this is the perfect place. It’s almost like a mini world,” says Dr. Schadt.
Mount Sinai may have extraordinary resources, but Dr. Schadt and the Institute also collaborate with other institutions, including Stanford, Harvard, MIT, Yale, UCLA, and others. Further, he mentions that, “We are seeking to partner with different pharmaceutical and biotech firms. Such companies are eager to partner and leverage our disease models and clinical setting to carry out human proof of concept studies.”
Before Mount Sinai
The network of events that came together to bring Dr. Schadt to Mount Sinai itself involves considerable complexity. As a child in Stevensville, Michigan, population 1,000, he hardly seemed destined for a scientific career. His stepfather was a beautician when the future scientist was a boy, and the family members were all creationists.
Education wasn’t valued, and Dr. Schadt was an indifferent student. It wasn’t until he joined the Air Force and took a battery of aptitude tests that anyone realized his potential as a mathematician. The Air Force provided him with a scholarship to Cal Poly, and in this new and exciting world, he majored in computer science and applied math, followed by studying for a doctorate in pure math at UC Davis.
Although he finished the course work and exams for a Ph.D. at Davis, pure math became less and less satisfying. Wanting to do something that would more directly help people, he enrolled as a Ph.D. candidate in UCLA’s biomathematics program. That eventually led to a career at Merck.
While at Merck, he and some of his similarly minded colleagues came to understand that drugs targeting one gene may marginally solve one problem, e.g., diabetes, but as the drug perturbed the network that the target gene operated in, it might well cause additional problems.
For instance, in the case of diabetes, one promising drug turned out to help mitigate the effects of a gene for diabetes, but it also created additional risks for obesity and cardiovascular disease. Further, targeting single genes was not enough to effect the right type of change over the vast molecular networks that operate within any given individual.
Instead, Dr. Schadt believed that to effectively treat most common disorders, networks of genes must be targeted. Merck, however, balked at adopting his network approach to drug development and, in 2009, he and his fellow Merck colleague, Stephen Friend, left to start Sage Bionetworks. Then came the offer to head Mount Sinai’s Icahn Institute for Genomics and Multiscale Biology.
Reluctant Transplant
Dr. Schadt moved from California to New York in 2011. Although he now finds Mount Sinai and New York a perfect location for himself and his family, he was reluctant to leave California. His family loved the Golden State and no one wanted to leave.
However, professionally, he realized that Mount Sinai would be perfect for him, considering the leadership, the vision, and the financing that were available. When he added to that the opportunity to head an institute that included a major genetics research enterprise embedded in a large clinical setting, it wasn’t really a choice.
One day he told his family, “Too bad, we’re coming to New York!” To the surprise of all, they ended up loving the city and enjoying life on the East Coast.
Dr. Schadt has many reasons to be pleased with his new environment, but one is the ethic of philanthropy that pervades New York. “This city has some of the most philanthropic people I’ve ever met,” he marvels. “Carl Icahn, for example, has given $200 million to Mount Sinai. The impact of a gift like this is without compare.”
Eric Schadt may be a scientist who is fascinated by the mathematical tools and techniques of big businessmen and money managers, but fortunately for science and genetics, they—Carl Icahn is an example—are fascinated and supportive of him as well.
With the vast new computational and mathematical tools now available at Mount Sinai, plus the unprecedented financial resources, Dr. Schadt and his institute are on the road to something transformational, something that can touch the lives of every person: faster, safer, more effective healthcare.
To see a list of Dr. Schadt’s top five genomic predictions, click here.
Mitzi Perdue, GEN’s corresponding editor, holds degrees from Harvard and George Washington University. She has authored more than 1,600 newspaper and magazine articles on science R&D and clinical medical applications, as well as on food, agriculture, and the environment. Perdue has a strong understanding of complex scientific and mathematical concepts. For 22 years, she was a syndicated columnist for the Scripps Howard News Service and before that, California’s Capitol News. Perdue is also the author of the newsletter from the professional association, Academy of Women’s Health. She has produced and hosted more than 400 interview shows, often in conjunction with scientists at the University of California at Davis. She is a former Commissioner for the U.S. National Commission on Libraries and Information Science and a former Trustee for the National Health Museum.



