December 1, 2006 (Vol. 26, No. 21)
Analysis of Custom Interactions Data in the MetaCore Environment
Over the last few years, we have witnessed the rapid evolution of software tools for functional analysis of microarray gene expression, proteomics, metabolomics and other omics data. Integrated data-mining platforms, such as MetaCore and MetaDrug (GeneGo; www.genego.com), combine manually curated databases of protein interactions and pathways with sophisticated network-analysis tools, and are becoming the mainstream in drug discovery and life science research.
The networks and pathways are generated from subsets of high-fidelity binary protein-protein, protein-compound, and protein-DNA interactions collected in the database, followed by statistical analysis of their relevance to specific functional processes, diseases, and toxic categories. The subsets of affected proteins, genes, and metabolites are defined in the omics experiments, which typically deal with human tissue samples in different therapeutic areas: human cell lines and mouse and rat data in toxicogenomics and drug response.
Reliance on the backbone of high-fidelity interactions extracted from full-text, small-experiment articles is key in analysis of inherently error-prone omics data sets, which otherwise are poorly comparable. However, high quality comes at a cost of restricting the analysis to the subset of mammalian proteins (genes), whose function is experimentally proven and for which interactions are published in small-experiment literature. Such information is not yet available for almost half of human proteins (defined as mRNAs).
On the other hand, omics experiments, such as yeast-to-hybrid assays, co-expression, pull-down immunoprecipitation, ChiP/Chip, microRNA assays, and high-content screening, are a rich source of putative physical interactions and functional associations between uncharacterized and known proteins as well as novel interactions between known proteins. Large pools of such custom interactions are accumulated at drug companies and in the public domain. Integration, alignment, and prioritization of potentially IP-rich but low-trust omics data with high-trust small-experiment data is the subject of active research in functional data-mining and network analysis.
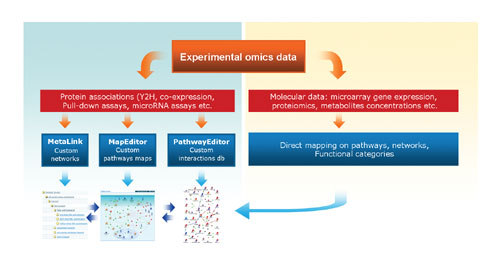
Here, we present three novel tools, which help analyze custom interactions within MetaCore and MetaDrug.
The general schema of integration and analysis of custom interactions is shown in Figure 1. The interactions can be visualized on networks using MetaLink™ and on static, interactive, canonical pathway maps using MapEditor and/or added to the underlying MetaCore interactions database using Pathway Editor. In all cases, the interactions themselves, as well as the derivative products (networks, pathway maps, and report tables), are accessible in secured user accounts and can be shared between individuals or a group of users.
Typically, interactions are not the only type of data analyzed by users, they accompany different molecular datasets such as the level of gene expression in microarray experiments, MS proteomics, or metabolomics concentration data. Therefore, the interaction tools work in sync with the MetaCore modules for visualization and statistical analysis of molecular data.

Figure 1
Customized Networks
MetaLink enables import of custom interactions into MetaCore, generation of networks with custom links as input edges using MetaCore algorithms and content, mapping molecular data on custom networks, and prioritization depending on interaction weights. The interactions are parsed into MetaCore in text or Excel format as tables with two columns of gene or protein IDs, which show association in the experiment and an optional table with the relative weights for each interaction specified by the user.
Usually, the weights reflect relative interaction trust levels associated with the experimental procedure used (yeast-two-hybrid, immunoassay, co-expression) or with the importance of the interacting proteins in the condition. Network-building algorithms in MetaCore take into account the edge’s weight and therefore the custom-defined weights will affect the topology of the resulting networks and their relevance to the input data.
An example of a customized network is shown on Figure 2. The associations data was generated for a set of cancer-related proteins in a simulation test, followed by generation of a network using the direct interactions (DI) algorithm. Previously published breast cancer SAGE gene-expression data was mapped on the resulting network.
The network represents a graph with four types of interactions, marked in different colors. Custom interactions with no corresponding interactions in the MetaCore database are visualized as pink edges. The edges that correspond to interactions both in the custom file and MetaCore’s database are marked blue, and MetaCore interactions not in the custom file but used by MetaCore network algorithms are marked with standard MetaCore edge colors, red, or green. One more type of edge (marked yellow) is for custom interactions present in the MetaCore database but not used for network generation by default (as low-trust interactions).
Most importantly, the gene-expression data is mapped on the custom network and visualized as red (up-regulation) and blue (down-regulation) circles with color intensity corresponding to the relative expression level. The new networks can be saved in the user’s account, and the interaction files and networks can be shared within a user group or among individual users. The gene content of custom networks can be exported as a list for additional analysis in third-party programs such as DecisionSite (Spotfire), Genespring (Agilent), Expressionist (GeneData), and Resolver (Rosetta Biosoftware).

Figure 2
Map Editor
MapEditor is a Java module that enables custom editing of canonical pathway maps available in MetaCore, conversion of networks into map visualizations, and drawing of regulatory and metabolic maps from scratch. A user may choose the objects for his/her maps from MetaCore’s content of genes, proteins, compounds, and interactions, or introduce new objects. Custom maps can then be securely published (added to MetaCore’s standard map collection on the customer’s server), used as a template for mapping experimental data, and/or saved and shared with colleagues.
A user can focus and organize his/her wet lab or dry lab research around a set of interactive maps created in MapEditor and linked to MetaCore. The custom set of maps can then be published and made available in MetaCore as a set of canonical signaling and metabolic maps. The user is then able to work with a vast body of high-throughput data (for instance, disease-specific microarray expression experiments) available internally or in public domain databases.
The data can be parsed in MetaCore and placed on custom and standard maps. The maps can be aligned according to relevance to imported data, and the gene content from multiple maps exported or explored further. Such capability of meta-analysis could substantially expand the focus of research of any biologist or medicinal chemist, regardless of access to high-throughput data.
An example of a MapEditor application is shown in Figure 3. A pharmaceutical MetaCore user studied how the apoptosis mechanism can be triggered in tumor compartments where p53 is inactivated due to mutation or deletion, critical information for cancer treatment. The initial set of proteins related to p53-independent G1/S DNA damage checkpoint was assembled from the published human Reactome and used to build a network.
Text-mining tools were applied to add interactions extracted from current literature articles using keywords such as p53-independent apoptosis. The combined list of protein IDs was then uploaded into MetaCore to construct a molecular network, which defines the relationship of these proteins. In MapEditor, the network was converted into MetaCore’s map format and compared with MetaCore’s internal p53-dependent apoptosis map to generate a global network reflecting apoptosis events mediated by the p53 protein family.
Microarray gene-expression profiles from three breast cancer samples from a previously published study were mapped on the network and the custom map. Importantly, this custom map was shared confidentially within a customer’s user group accessing their internal MetaCore server for feedback and stimulated discussion.

Figure 3
Ongoing Annotation Projects
PathwayEditor, which is used by GeneGo’s annotation team for entering and editing interactions in MetaCore (Figure 4), is designed primarily for larger customers with ongoing internal annotation projects. The PathwayEditor represents a series of JavaScript tables with fields detailing the mechanism and type of interaction, experimental procedures as the interaction evidence, trust level, the participating proteins, isoforms and complexes, literature references, and other relevant data.
Unlike MetaLink and MapEditor, the interactions introduced by using PathwayEditor stay permanently in the customer’s version of MetaCore. The information describing novel objects (proteins, genes, interactions, compounds) will be parsed into multiple tables in the MetaCore database and will become eligible for search queries. The new objects will be included as nodes and edges in network generation in the custom version of MetaCore.
We believe that the suite of customization tools described in this article helps to integrate high-confidence small-experiment protein interactions with noisy but information-rich omics-generated associations and molecular data within the same data-mining platform. All three products are add-ons for MetaCore, integrated with the platform’s analytical and visualization tools.
The corresponding molecular data can be mapped and prioritized on the objects introduced into networks and pathways with the help of new tools, therefore completing integration of multiple types of omics data.

Figure 4
Yuri Nikolsky is CEO, Julie Bryant is vp of business development and sales, and Tatiana Nikolskaya is founder, CSO, and president at GeneGo. Web: www.genego.com. E-mail: [email protected].