December 1, 2011 (Vol. 31, No. 21)
Vicki Glaser Writer GEN
Use of NGS in Medicine Contributing to Improvements in Human Health
A plenary debate, entitled “Current and Emerging Sequencing Technologies: Changing the Practice of Medical Genetics,” provoked strong and divergent opinions at the combined American Society of Human Genetics (ASHG) and International Congress of Human Genetics (ICHG) conference held recently in Montreal.
Han Brunner, M.D., Ph.D., a clinical geneticist and professor of human genetics at University Hospital and Radboud University, moderated the debate, which in the end generated more questions than answers, highlighting the strong opinions and emotions surrounding the role of genome sequencing in medicine today and into the future.
The panelists represented a range of viewpoints, expressing the perspectives of industry, researchers, clinicians, and patient advocates. They included Joris Veltman, Ph.D., associate professor, department of human genetics, Radboud University; Radoje Drmanac, Ph.D., co-founder and CSO at Complete Genomics, Ségolène Aymé, a medical geneticist and epidemiologist, director of research at INSERM and coordinator of the Orphanet platform for rare disease registries; Louanne Hudgins, M.D., professor and chief, division of medical genetics in the department of pediatrics at Stanford University School of Medicine; Michael Hayden, Ph.D., director and senior scientist, Centre for Molecular Medicine and Therapeutics, and University Killam professor, department of medical genetics, University of British Columbia; and Ming Qi, Ph.D., director and professor, Zhejiang University Center for Genetic and Genomic Medicine, China.
The panelists each had an opportunity to respond to four statements, and then to discuss and debate each others’ responses: 1) Targeted sequencing will remain the norm for diagnostic medical genetics because whole exome and whole-genome sequencing (WGS) will yield an excess of information that is useless, counterproductive, and possibly damaging to the patient. 2) Personal genome sequencing creates an unacceptable risk to the privacy of people. 3) Cytogenetics will cease to be. Sequencing is the only future technology in diagnostic labs. 4) Personal genomes will be incorporated in the standard of care for all medicine. Therefore, medical genetics will disappear as a separate medical specialty.
Han Brunner summed up the main challenge implicit in all of these statements, asking the panelists, “How do we take this beast of a new technology and use it for the advantage of our patients?” And that, above all else—how to use WGS to improve human health—was the facet on which the panelists could all agree, that ultimately, the use of next-generation sequencing (NGS) technology in clinical medicine had to benefit patients and their families.
Notwithstanding that the ability to do targeted DNA sequencing, exome sequencing, or whole-genome sequencing accurately and cost effectively is a remarkable scientific achievement and is here today, or that the data generated has incalculable value from a scientific and research perspective, are we ready to share this information with clinicians and patients, do we really know what it means in terms of predicting inherited disease risk, diagnosing disease predisposition, providing reproductive counseling, or managing disease, and do we know how to interpret and apply sequence information to impact prognosis, treatment, or quality of life?
The areas of consensus were quite clear, the points of disagreement less so, as the panelists debated: whether genome sequencing technology should be implemented simply because it is available; whether it is immoral not to gather the most information you possibly can about a patient’s disease; who “owns” and has a responsibility to store, protect, and reanalyze individuals’ genomic data; what do patients want to know; and where is the line (and how do you keep it from blurring) between clinical research and clinical medicine and when is it appropriate to cross the line from applying NGS information for scientific discovery to using it to guide clinical decision making; and how do we maintain a clear distinction between disease-related sequencing and screening, and should clinicians handle incidental findings.

Next-generation sequencing technology is rapidly moving toward the clinical diagnostic arena while remaining a critical basic research tool. [iStockphoto]
Where We Can Agree
Several main areas of consensus emerged from the discussions, including: the directive to proceed carefully and, as in all aspects of medicine, first do no harm; to continue to emphasize hope for what NGS can provide, but without all the hype; and to acknowledge that whole-genome sequencing is here now and is here to stay. Overall, the panelists agreed that, with the exception of diagnosing and providing genetic counseling on Mendelian diseases, sequence data is “not ready for prime time” and it is far too early to share the information with patients, and perhaps even with clinicians.
Exome Sequencing Finds Sweet Spot: At least in terms of sharing information, the panel said, the gold standard remained targeted sequencing for identification of specific genetic diseases. But somewhere between targeted sequencing and likely uninformative whole sequence data lies exome sequencing, a technique that selectively targets the most functionally relevant DNA sequences that encode proteins. You can get the scoop on how scientists are leveraging exome sequencing here.
Three overarching and urgent needs emerged from the debate: unified strategies for storing and interpreting genomic sequence data; mechanisms to protect patient privacy and prevent misuse of genetic data to stigmatize or discriminate against individuals; and the need for education—for physicians, medical geneticists, genetic counselors, and patients—on how to present and share sequence data and what it may or may not mean.
There was general support for the creation of an international, curated genotype/phenotype database to help accelerate and standardize data interpretation and understand differences based on ethnicity, and for an international, multidisciplinary body of experts that would establish underlying principles, monitor the field as it advances and evolves, help determine what filters are needed between sequence information and clinicians/patients, and devise strategies to protect individual privacy.
There was no consensus or definitive answers to several issues raised, such as how to pay for testing and what it will take to convince payers that whole-genome sequencing is cost-effective; what constitutes true informed consent; and who should have access to a patient’s sequence information.
The panelists challenged each other to delineate the point at which DNA sequence data has clinical utility—when it is “actionable” and how to define what actionable even means. Does it have to guide treatment decisions for a patient with a diagnosed disorder or is it actionable if it can simply improve quality of life and perhaps impact a person’s risk of developing a disease?
For example, would it be valuable to be able to tell patients they have a genetic trait associated with an increased risk of high cholesterol if it might encourage them to change their lifestyle and behavior to modulate that risk?
Another complex question arose: for WGS data collected now and stored for future use, who is responsible for maintaining, updating, and re-analyzing the data as new knowledge becomes available? And is there, in fact, a moral obligation to reanalyze the information and report the results to patients?
Where clear differences of opinion emerged was in discussions comparing the short-term and long-term value of targeted genomic sequencing, whole-genome sequencing, and exome sequencing. Should the focus be on collecting WGS data because there is more of it and because we can or is it more appropriate and cost-effective to redo targeted sequencing as needed?
A Difference of Opinion
Will targeted sequencing remain the standard for diagnostics for the near future? An emphatic, “No,” said Dr. Drmanac. “Targeted sequencing does not give you the right [or complete] answers,” he insisted, making his case for whole-genome sequencing. His company, Complete Genomics is in the business of sequencing whole genomes. He gave three main reasons to support a move toward WGS: genes and regulatory networks are so intertwined and interdependent that contextual interpretation of the sequence data is only possible with whole genome data; all human variation cannot be put on a chip; and we should celebrate the ability to sequence whole genomes.
According to Dr. Drmanac, the technologies available today are scalable and he predicted that in three to four years these methods will make it possible to sequence one million genomes per year.
Regarding the fourth statement, Dr. Drmanac commented that personal genomics is “unstoppable.” It will not displace medical genetics as a field; both medical genomics and medical genetics are needed, as are tests for somatic mutations.
Dr. Drmanac said that WGS information should be acquired as early as possible in a person’s life, with testing done on newborns and prior to embryo selection and implantation in IVF procedures. While the field should focus on what is actionable, that should not limit WGS. He encouraged his colleagues to find all sequence variants in a genome but only report those known to be disease-related.
Currently, Complete Genomics offers sequencing services and provides data only to researchers, who must have IRB consent from patients. In 2012, the company plans to seek CLIA certification, which will enable it to provide data directly to physicians.
In Dr. Hayden’s view, “We have oversold the concept of the genomic revolution.” The promise of the Human Genome Project and genomics has not been fulfilled for the general public, he added. “Don’t oversell the promise of whole-genome sequencing,” noting the excessive hype already surrounding genome-wide association studies (GWAS) and the promise that they will lead to new drug targets and therapies.
In response to statement 1, he questioned what we would do with the information from WGS. There is a “massive information gap” and educational gap, in addition to an absence of clear stakeholders and no reimbursement strategies in place. “Technology is not the problem; delivery is the problem”—and even how to define delivery is a problem. “We are not ready for WGS; for research use, yes, but for patients, no.”
Dr. Hudgins, focused on statement 4 and responded both yes and no. Yes, she feels that personal genomes will be incorporated in the standard of medical care, but how, when, and where are not clear. She vehemently disagreed with the idea that the availability of genomic information will replace medical genetics though.
Personal genomics will change the face of diagnostics, she contended, and instead of relying on phenotype and test results alone, genotype information will be used to support a diagnosis. Dr. Hudgins warned, however, that even those trained in medical genetics are ill-prepared to make accurate predictions based on genetic information related to inheritance of and predisposition to complex disorders. “GWAS are not the whole story,” she added, emphasizing the importance of epigenetics and somatic mutations.
Dr. Veltman based his views on his group’s experience sequencing 500 exomes for clinical research purposes, not patient care, to identify Mendelian disease genes with de novo and inherited mutations related to intellectual disability and blindness. He focused on the issue of protecting patient privacy and safeguarding sequence data. He does not believe that targeted sequencing should be the norm, because one should not assume that a problem lies in only certain genes. Although he “believes strongly in the importance of technologies that have no bias,” he pointed out that “we can implement exome sequencing now.”
Aymé represented the voice of the patients, whose needs, she asserted, should be the focus of the discussion. Those needs “have not changed simply because the technology has advanced,” she said. Patients want to know whether they have a familial disease and whether there is a treatment. “People want to know about their disease, not their disease risk.”
Genomic information should be used only if it would modify the prognosis, concluded Aymé. “I am in favor of developing the scientific aspect of sequencing,” she said, but cautioned against saying that it will improve the quality of care or exposing patients to sequence information unless there is a specific medical reason for doing so.
Faster Methods for Nucleic Acid Detection, Amplification, and Sequencing Showcased at ASHG/ICHG Conference
OpGen introduced Genome-Builder software for sequence assembly. The company presented the results of initial studies using Genome-Builder done in collaboration with the Sanger Institute. OpGen designed the software to work together with its Argus Whole Genome Mapping System to combine the results of whole-genome mapping with partially assembled sequences to detect structural rearrangements.
Affymetrix launched version 2.0 of the GeneChip® miRNA Array, designed to detect short mature active miRNAs, longer inactive precursor miRNAs, small nuclear RNAs (snoRNAs), and small Cajal body specific RNAs (scaRNAs), representing small noncoding RNAs involved in cellular post-transcriptional regulation.
The company’s Gene 1.1 ST Array Strips for the GeneAtlas® System capture whole-genome expression at the gene and transcript level for a variety of model and applied research organisms. Affymetrix released its Axiom® Genome-Wide Pan-African Array, designed to maximize genomic coverage of common and rare alleles in populations of African ancestry.
RainDance Technologies launched the ThunderStorm™ system for next-generation targeted sequencing. The system can automatically process up to 96 samples and access up to eight primer panels in performing microdroplet single molecular PCR amplification.
ImmunoSEQ is using next-generation sequencing technology to perform immunoprofiling of adaptive immune receptors, and is currently developing initial clinical applications focused primarily in oncology for use in monitoring residual disease.
Diagenode showcased two instruments that it brought to the market this year: the Bioruptor® sonication device for DNA and chromatin shearing, with simultaneous processing of up to 12 (200 bp) samples in 23 minutes, parallel processing of 48 samples/run, controlled DNA fragment size range, and scalability from microliter to milliliter quantities; and its SX-8G automated systems for epigenetic assays.
The QuantStudio™ 12K Flex real-time qPCR system, new from Applied Biosystems (a Life Technologies company), offers the capability to scale experiments from a 96-well plate format up to a 3,072-reaction OpenArray plate. The system can process up to four plates in parallel, or more than 12,000 samples per run in about four hours.
The instrument contains five interchangeable thermal cycling blocks. Users can switch between qPCR and digital PCR modes OpenArray, Taqman Array Card, 384-well, and standard or Fast 96-well blocks. The accompanying AccuFill™ system facilitates sample loading.
Roche plans to introduce version 3 of its NimbleGen Sequence Capture genomic-enrichment technology, with refinements that the firm says will improve target exon capture by about 50% and will simplify multiplexing by allowing researchers to pool libraries before the sequence capture step.
This month, Roche launches the LightCycler® Nano real-time PCR system in North America, in which reactions take place in reaction tubes on plastic strips, with eight tubes/strip. Also in development at Roche is an upgrade package for the 454 Life Sciences’ Genome Sequencer FLX system. The new FLX Plus will offer longer read lengths for NGS—up to 1,000 base pairs.
Agilent Technologies showcased a high-throughput version of the 2100 Bioanalyzer called the 2200 TapeStation, a benchtop gel-based capillary electrophoresis system that can analyze 16 samples/run at a rate of about 1 minute/sample.
Thermo Scientific featured the PikoReal real-time PCR system, which is available in two configurations for use with 24-well plates and 8-well strips, or with 96-well plates. Benefits of the instrument, according to Thermo, are its small footprint, low energy usage, and reduced plastic and reagent consumption.
Fluidigm launched an expanded version of the Access Array™ integrated fluidic circuit-based target-enrichment technology for targeted resequencing applications with NGS. The new version makes it possible to multiplex up to 10x amplicons across 48 samples, or 480 amplicons/chip.
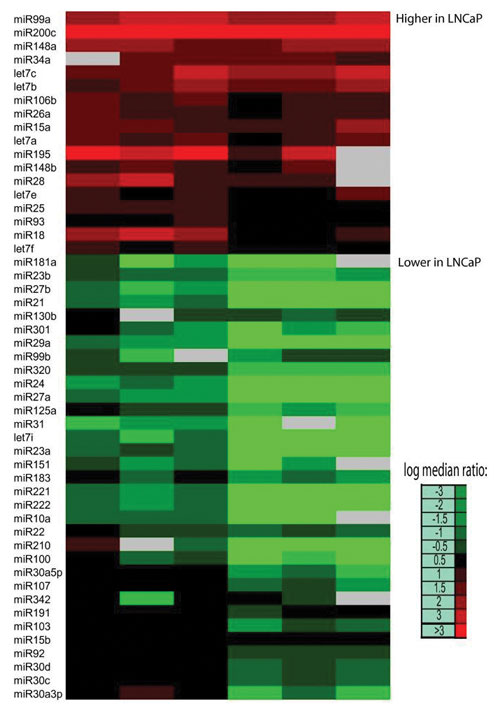
Microarray comparison of microRNA expression in PC3 and LNCaP cells. Expression is displayed as higher (red) or lower (green). [Affymetrix]
Rubicon Genomics introduced the NovaPLEX™ Sheared DNA Prep Kit, which uses ligation-based library synthesis and library amplification technology for preparing double-stranded DNA for the Illumina NGS platform. It utilizes a single-tube workflow that does not require intermediate purification and can accommodate sample input down to 50 picograms.
The HaloPlex™ PCR Reagent Kit from Halo Genomics is used to prepare PCR products for targeted resequencing applications without the need for dedicated instrumentation. It can amplify 10–2,000 exons in one tube and allows for sample multiplexing.
Complete Genomics introduced its new cancer-specific whole-genome sequencing service. The results include somatic and germline variations including SNPs, insertions/deletions, structural variations, copy-number changes, and mobile element insertions.
Illumina introduced three workflow enhancements to support its MiSeq personal sequencer system: the BaseSpace™ NGS cloud-computing informatics environment; TruSeq custom amplicon kits for preparing and enriching up to 384 amplicons per sample and up to 96 samples per plate in parallel in less than eight hours; and Nextera™ DNA sample-preparation kits.
Pacific Biosciences has partnered with Cycle Computing to optimize the PacBio RS SMRT Analysis software for cloud-based applications. The company plans to offer a beta version of the new software suite with the next major upgrade of its RS sequencing system.
New from Tecan are the HydroSpeed™ plate washer in 96- and 384-well plate formats and the Infinite® M1000 quad4 monochromators™-based multimode reader.
Qiagen has added preprogrammed protocols to its QIAxcel® Advanced automated capillary electrophoresis system to help standardize DNA and RNA detection.
PerkinElmer added a multiple myeloma chip to the OncoChip™ microarray family, designed to identify chromosomal abnormalities associated with hematological cancers. The company also updated its Signature Precision Panel™ prenatal test.
NanoString Technologies introduced the second generation nCounter® Analysis System, a digital detection and counting system for copy number variation, gene expression, and miRNA profiling.
DNAnexus entered a partnership with Google to provide access to DNA sequence data stored in the public Sequence Read Archive database, with a free web-based search interface and support from Google Cloud Storage.
DNA Genotek, acquired by OraSure Technologies in August, initiated a grant program aimed at cancer, personalized medicine, and infectious disease research. The company will review research proposals for innovative uses of its saliva-based collection and stabilization products for genomic/genetic studies.
NextGENe NGS analytical software from SoftGenetics is compatible with the Ion Torrent PGM™, Roche Genome Sequencer FLX, Illumina Genome Analyzer, and Life Technologies SOLiD™ sequencing systems. Its features include a single annotated data review screen, a Windows interface, a Mutation Report hyperlinked to the graphical NextGENe browser and dbSNP database, and a variety of filtering options. n
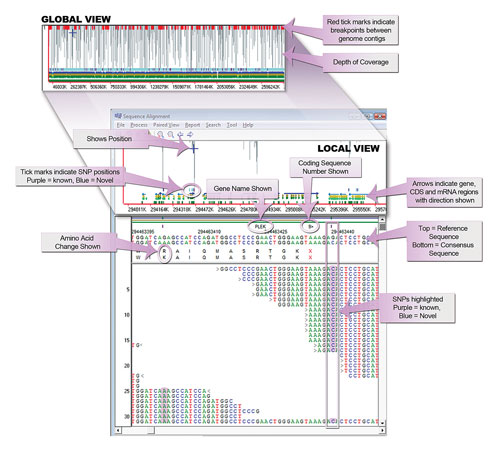
NextGENe analysis browser provides an interactive review of annotated analysis results in a single view. The browser provides a global view of the entire analysis (top panel), whole human genome pictured here, indicating coverage depth across the analysis, break points between contigs, red ticks, as well as linked localized view, which includes color-coded indication of found variants, CDS and mRNA regions, reference and consensus sequence nucleotide and amino acid sequences, gene name and coding sequence number, as well as a view of actual base call by aligned read.