June 15, 2009 (Vol. 29, No. 12)
Anthony L. Shrout
Template-Directed Assembly Has Role in Assessing Activity, Regulation, and Recognition
Many diseases are initiated from molecular events that take place at the cell membrane level. The membrane plays a critical role in cell communication and is the launching point for many vital signaling processes that occur within and between cells.
Membrane-associated proteins either span the membrane, or are closely associated with the surface through domain interaction or through a number of posttranslational modifications that allow them to target the membrane.
Many of these proteins present a unique set of challenges to the researcher. Because of the inherent problems associated with designing assays for this class of proteins, researchers are forced to work with only the small cytoplasmic fragments of receptor tyrosine kinase (RTKs) domains. While these soluble fragments are simple to purify, they do not always exhibit proper biological function or normal activity because they have been removed from the fluidic environment of the membrane.
The membrane surface provides a unique organizing influence that allows complex signaling units, often composed of many different proteins, to form and carry out their natural function in a dynamic environment. When removed from the membrane environment, these proteins often exhibit low activity and frequently require the addition of nonphysiological counterions or other molecular crowding agents just to recapitulate some of their activity.
Protein Attachment Technologies has developed a method to facilitate the interrogation of membrane-associated proteins. Template-directed assembly (TDA) makes use of the ubiquitous histidine tag (His-tag) that has gained popularity in protein purification and in structural studies over the last two decades.
The TDA platform is a stable liposome that is decorated with NiNTA headgroups. When users combine His-tagged receptor fragments with TDA’s synthetic liposomes, proper enzymatic function is restored and the ability for the proteins to form multiprotein complexes is obtained. This technology was specifically designed for proteins that normally reside at or near the membrane surface—soluble cytoplasmic kinases anchored to TDA nanospheres show no improvement of activity, as expected.
While TDA is simple in principle, there are procedural and engineering concerns that must be considered. For example, TDA nanospheres are sensitive to EDTA and large concentrations of detergents. Additionally, membrane-associated proteins are frequently sensitive to changes in their juxtamembrane domains and may need to be engineered specifically to function on TDA nanospheres in addition to being N-terminally His-tagged for proper orientation. Thus, some care must be taken when designing proteins and reagents for use with TDA.
The end result, however, is a more biologically relevant system—more complete cytoplasmic domains that interact as they do in the cell, lower concentrations of ions, and proteins that are placed back into the membrane environment. Figure 1 depicts how TDA enables protein assembly.
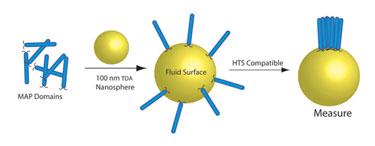
Figure 1. Soluble membrane-associated proteins (MAPs) assembled (in solution) and on TDA nanospheres where they then assemble and interact as they would in a natural membrane environment.
Activity and Enzymatic Regulation
TDA nanospheres are fluid in nature and, therefore, allow anchored proteins to migrate and interact as they would in a natural cell membrane. Studies have demonstrated increases of up to ~50-fold, in overall substrate phosphorylation by insulin receptor kinase domains, as well as increases in autophosphorylation (Figure 2, left) with TDA. Autophosphorylation activity is typically not observed to a measurable extent in conventional solution assays; the results obtained with TDA demonstrate that the assembled proteins are interacting in a biologically relevant fashion, i.e., undergoing transphosphorylation events.
TDA nanospheres, however, do not simply increase activity. The rightmost portion of Figure 2 represents experiments using template-assembled Tie2 kinase domains. This protein has a known role in angiogenesis and is linked to certain types of cancers. Atypical of RTK function, when Tie2 undergoes transphosphorylation the rate of substrate phosphorylation decreases; this behavior occurs only on the assembled protein as demonstrated in Figure 2 (right, + Template).
This interpretation leads to the conclusion that solution Tie2 experiments (- Template) demonstrate aberrant kinase behavior indicative of a protein that is not properly functioning compared to the in vivo assay.
These results agree with recent crystal structure data as well as physiological studies of Tie2. While encouraging, these results represent only the basic hallmarks of properly functioning proteins. Further detail is needed to fully demonstrate the importance of the membrane environment and the control it exerts on the proteins that reside there.
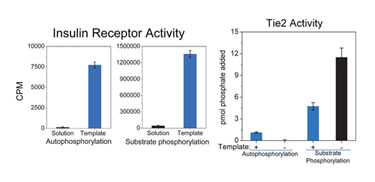
Figure 2. Increased activity of insulin receptor kinase domains (phosphorylation of substrate and autophosphorylation) and demonstrable regulation of Tie2 domains result from proper assembly using TDA nanospheres (template).
Altered Substrate Recognition
TDA provides the environment needed for increased activity and proper regulation of many RTK domains. Data indicates that proteins assembled on TDA nanospheres adopt different conformations, leading to different substrate recognition profiles. In a recent collaboration with Molecular Devices (MD; an MDS Analytical Technologies company), TrkB tyrosine kinase domain was analyzed with and without TDA nanospheres on MD’s SubstrateFinder assay platform.
The substrate profile was shown to be remarkably different with the assembled TrkB versus TrkB in the conventional solution assay—nine new substrates were identified using template-assembled TrkB, while eight substrates recognized by untemplated enzyme were not recognized by template-assembled TrkB. This data reveals a common theme among proteins that are normally associated with a membrane, namely that it is critical to place these proteins back into the environment in which they evolved to function.
Complex Assembly Using TDA
TDA technology was first developed by assembling a four-protein complex from an E. coli chemosensory system. The chemosensory system, consisting of a receptor, kinase, response regulator, and adaptor protein, provided a model system to explore the usefulness of template-directed assembly.
Using TDA, it was demonstrated that one could assemble multiple proteins, the base of which was a fragment of the chemoreceptor Tar; this is analogous to displaying IGF-1R on the surface and assembling IGF-1R:IRS1 complexes. In solution these four proteins did not assemble, and thus, exhibited no activity. When templated, however, the proteins formed functional complexes and exhibited in vivo activation levels and function.
The study of heteroreceptor interactions and subsequent regulation was also made possible by TDA. This data, given the similarities between bacterial and human-receptor signaling pathways, suggested that similar complexes could be assembled using human proteins. Figure 3 depicts how a human signaling-pathway system can be constructed on TDA nanospheres. This use of TDA facilitates screening by allowing for simpler experiments using multiple purified components.
Membrane-associated proteins represent a large, yet underexploited group of drug targets. Because they have been removed from their natural membrane environment, for ease of purification and structural studies, they become less active and behave sub-optimally in standard assays. TDA provides a novel way to assemble these proteins, leading to proper function and activity, thereby providing an improved way to discover active inhibitors.
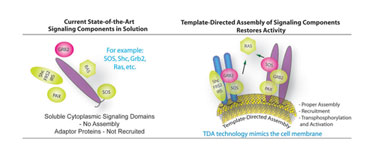
Figure 3. Current assays involve the interrogation of RTK domains only.
Anthony L. Shrout, Ph.D. ([email protected]), is CSO at P.A. Technologies. Web: www.patechllc.com.







