March 1, 2011 (Vol. 31, No. 5)
Vicki Glaser Writer GEN
Advanced Technological Approach Generates Genomic Data Better, Faster, and Cheaper
In the next-gen sequencing (NGS) arena, the focus over the past several years has been on technological advances, moving from second-generation to third-generation sequencing strategies and producing research instruments capable of delivering whole-genome sequences in parallel at increasing speed. More recently, as read lengths and coverage continue to increase, throughputs rise, and costs decline, the expanding range of applications of NGS has taken center stage.
Concurrently, broader accessibility and affordability of NGS and its promise in the clinical arena have captured the spotlight with the emergence of two new “personal” sequencing systems, opening the door to sequence-based diagnostic and prognostic applications, tumor profiling, treatment selection, and patient stratification for clinical trials. The ability of the NGS technology available today to deliver the goods is evident in the steady stream of whole-genome sequences being reported across microbial, plant, and animal species.
Genomic Health announced results from its next-gen sequencing-driven biomarker discovery program in breast cancer at the recent “Advances in Genome Biology and Technology” (AGBT) meeting. Based on sequencing of the whole human transcriptome in formalin-fixed paraffin-embedded (FFPE) tumor and normal breast tissue samples, the company found hundreds of differences in both coding and noncoding transcripts between the two sample populations. Genomic Health reported an association between specific genes and some non-coding RNAs and risk of breast cancer recurrence.
Cholera
As recently reported in the New England Journal of Medicine, a collaborative effort between researchers at Harvard Medical School and Pacific Biosciences produced the genome sequence for the strain of Vibrio cholerae responsible for the recent cholera epidemic in Haiti. The strain is related to a South Asian cholera variant and had not previously been documented in the Caribbean region or Latin America.
PacBio applied its single molecule real-time (SMRT™) DNA sequencing technology to decode two samples from the recent Haitian outbreak and three other strains of V. cholerae and compared them to DNA sequence information for 23 cholera strains available in public databases. Sequencing of the five sample genomes was completed in less than two days, demonstrating the potential for using NGS for rapid pathogen identification in outbreak situations.
PacBio is in the late stages of its limited production release (LPR) program, optimizing and upgrading the chemistry and software for its beta version RS system and performing validation studies in preparation for a first half of 2011 launch date for the commercial instrument.
Eric Schadt, Ph.D., CSO, presented the cholera study data at AGBT. He highlighted two key advantages of the RS system that have become evident during the LPR period: rapid sample turnaround and the value of long read lengths. Quick turnaround is especially important in the infectious disease space, noted Dr. Schadt. It will usher in “a new era in molecular epidemiology,” allowing a shift from phenotype-based to sequence-based determination of infectious strains.
Long read lengths help uncover large-scale structural variation such as copy-number variation or gene rearrangements, which may have a greater impact on function than SNPS or indels. Referring to the cholera example, Dr. Schadt said, “we were able to achieve 15x coverage in 90 minutes” and to identify large structural variations that contributed to unambiguous differentiation of the bacterial strains.
Dr. Schadt identified two main areas targeted for improvement: throughput and accuracy. The throughput of PacBio’s third-generation methodology does not yet match that of second-generation sequencing technology, and higher throughput will be needed to sequence larger mammalian genomes efficiently.
The first commercial RS system will contain a SMRT Cell with two sets of 75,000 zero-mode waveguides (ZMWs), which are nanometer-sized holes that function as a window for observing DNA polymerase-driven nucleic acid synthesis at the single molecule level. As the density of ZMWs increases, the throughput of a chip will increase.
In the short-term, hybrid assembly strategies that combine second-generation sequencing technology to generate highly accurate short reads and third-generation sequencing to achieve full coverage and assemble the short contigs into a complete genome will help overcome this limitation, in Dr. Schadt’s view. With regard to improving accuracy, PacBio’s beta instrument has an average raw sequence read accuracy of 86%, and the company is aiming for 85%–90% accuracy for the commercial system.
Applications in progress or in development at PacBio include identifying epigenetic modifications at full-genome scale, performing targeted resequencing of medically relevant genes to stratify patient populations, enabling direct transcriptome sequencing, and generating “disease weather maps” that can be used to identify and track changes in human pathogenic viruses present in the sewage system, water supply, and food supply of populated areas.

SMRT Cells are nanofabricated consumable substrates used in conjunction with the DNA Sequencing Kit for automated processing on the PacBio RS system. [Pacific Biosciences]
Bearing Fruit
Recent studies in the literature using Roche’s 454 sequencing technology include the discovery of a new human immunoglobulin (IGHV) gene and 16 new IGVH allelic variants published in the February 2011 issue of Immunogenetics. Additionally, a collaboration between 454, the University of Florida, the DOE Joint Genome Institute, and the Georgia Institute of Technology has borne fruit with completion of the first citrus genomes, including the sweet orange and the Clementine mandarin.
Researchers from the Munich Leukemia Laboratory presented on the use of 454’s Genome Sequence (GS) FLX and GS Junior systems for targeted resequencing of genomic regions associated with blood cancers at the annual meeting of the American Society of Hematology. The ability of long-read sequencing technology to detect multiple types of variation including point mutations, insertions/deletions, and structural variation made it possible to identify novel mutations in patient samples and to stratify patients into disease risk subtypes, which aids in diagnosis, prognosis, and detection of reemergence of resistant disease following therapy.
Soon to be introduced by 454 is new chemistry for the company’s GS FLX sequencing system that will enable longer reads of 700–800 base pairs and improve the quality of de novo genome assembly. The company also plans to launch a series of assays for application-specific workflows on both the GS FLX and GS Junior systems. The first assay will be primer sets for HLA genotyping, followed by assays targeting oncology, immunogenetics, and infectious disease applications.
Roche is also partnering with DNA Electronics to develop a low-cost high-throughput DNA sequencing system that will combine 454’s current pyrosequencing-based platform with DNA Electronics’ semiconductor technology. The system would rely on electrochemical rather than optical detection technology to monitor nucleotide incorporation during sequencing.

The GS Junior System offers a 10-hour run time for quick overnight sequencing and processing, according to Roche.
Getting Personal
MiSeq™, Illumina’s new low-cost personal sequencing system, leverages the same TruSeq™ sequencing chemistry that drives the company’s flagship HiSeq™ platform. MiSeq can take purified DNA and generate analyzed sequence data in about eight hours and, in just over a day’s time produce more than one gigabase at a cost of $400–$750 per run. Illumina plans to ship the first commercial MiSeq units this summer.
Key advantages of MiSeq are its fast turnaround time, ease of use, and simple sample prep, said David Bentley, Ph.D. Dr. Bentley, chief scientist at Illumina, envisions customers using the system for various types of applications: to check a small amount of sample before running it on HiSeq, to analyze large numbers of poor-quality DNA samples isolated from FFPE tissues, and to detect specific mutations in patient samples from clinical trial populations.
Illumina has doubled the yield from the HiSeq system to 600 gigabases, increasing the instrument’s capacity to 4–5 sequenced genomes in a 10-day run. Sufficient scale and throughput are now available to allow users to analyze and compare hundreds of samples, and these improvements are requiring users “to set up more sophisticated automation platforms,” said Dr. Bentley.
Illumina recently reached an internal milestone, completing a terabase run more than once. The company is nearing completion of work to validate HiSeq in its in-house CLIA sequencing laboratory in preparation for introducing the system into the clinical arena.
Although the capability of next-gen sequencing technology continues to outstrip available computing power, “these problems are tractable,” said Dr. Bentley, and the need for more powerful informatics tools is systematically being addressed. “We are drastically reducing the amount of storage space needed to store the same amount of information,” he explained, pointing to positive signs for the future including Cloud-based initiatives for centralizing data analysis and the development of increasingly sophisticated and diverse informatics strategies for analyzing sequence data and extracting new and focused information.
Dr. Bentley emphasized the need for software tools designed for “less expert users or experts in a different field.” As sequencing technology moves beyond the research laboratory, for example, it become increasingly important to design tools that will allow clinicians to extract and analyze the types of information most useful to them.

Life Technologies’ Ion Personal Genome Machine (PGM™) is based on Ion Torrent’s semiconductor sequencing chips that translate chemical signals into digital information. The 314 sequencing chip contains an array of 1.3 million wells; each is the site of an individual sequencing reaction. A pH change is detected when incorporation of a new base onto a growing DNA strand produces hydrogen ions.
The system is able to detect each base addition without the need for optics, a light source, or scanning detection technology. The 314 chip can yield at least 10 Mb of DNA sequence per run. From sample loading to raw data generation takes about two hours, with an additional hour for data analysis.
Life Technologies is rapidly scaling throughput of the PGM, and when the new 316 chip becomes available in 2Q2011 it will generate 10-fold more sequence per run, or 100 Mb. The company expects to maintain a pace of increasing the throughput of its sequencer by a factor of 10 about every 6 months.
The system’s housing and server will not change; users will only have to upgrade the chip and the software. The higher throughput chip will make it possible to sequence larger amplicon sets and whole microbial genomes on the PGM. Researchers have demonstrated the use of barcoding techniques to sequence multiple different samples on a single chip.
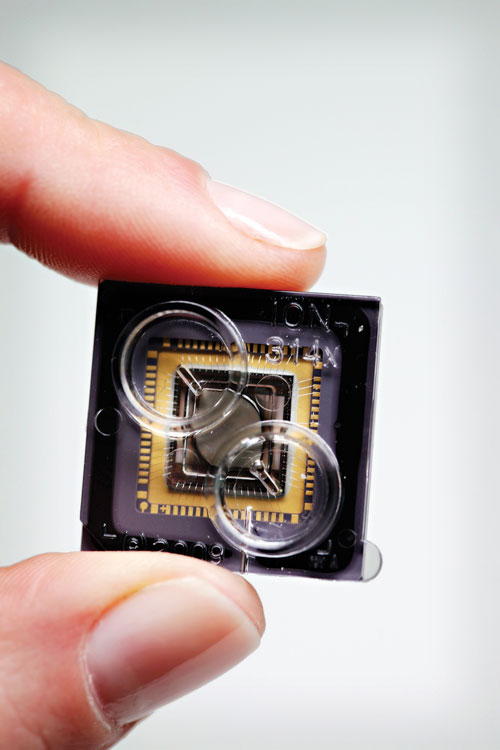
Ion Torrent’s Ion Personal Genome Machine is based on the company’s semiconductor sequencing chips, which translate chemical signals into digital information.
Going Public
Complete Genomics, which leverages its human genome sequencing capabilities through a service delivery platform, employs a sequencing method based on DNA nanoball (DNB™) arrays and combinatorial probe-anchor ligation read technology. The company optimized its sequencing technology specifically for the human genome and delivers to its customers annotated sequence data, identifying key sites of sequence variation.
It recently expanded its suite of analytical tools and announced the addition of copy number variation and structural variation results as part of its standard service. This enhancement has particular relevance for sequencing cancer genomes.
Complete Genomics became a public company in November 2010 and is moving forward with a number of large genome sequencing projects and partnerships. It recently received an order from the Institute for Systems Biology (ISB) to sequence 615 human genome samples for a study on neurodegenerative disease.
The company is also collaborating on a project with the NCI to sequence 100 genomes (50 tumor-normal pairs) as part of a pediatric cancer study to identify and validate somatic mutations associated with tumorigenesis. On successful completion of this initial phase, a follow-on NCI project will commence involving more than 514 tumor-normal pairs representing five different pediatric cancers.
The company recently made genome sequences generated for three members of a Yoruba family accessible to the global research community—they are part of the 1000 Genomes Project. At the AGBT meeting Complete Genomics announced plans to release 60 complete, high-coverage human genomes (including the Yoruba trio)—representing >12.2 terabases of mapped reads—to its newly established public genome repository.
Complete Genomics has the capacity to sequence as many as 400 complete human genomes per month, according to president and CEO Clifford Reid, Ph.D. It delivers the sequence data to customers on a hard drive or through the Cloud. Dr. Reid describes the company’s method as the “only unchained read technology in the industry. We can read bases in any order,” which contributes to higher accuracy and lower chemistry costs.
Confident that the size of the clinical sequencing market will one day surpass the research market, Dr. Reid described work under way at Complete Genomics to produce clinical quality sequencing data. Optimization of Complete Genomics’ long fragment read technology, which is now in development, will reportedly allow the company to reduce the error rate for its sequencing technology from 1 in 100,000 bases to 1 in 10 million bases, the latter equivalent to about 300 errors across an entire human genome.
The error detection/correction technology in development combines a DNA-engineering step upstream and an error correction step during data collection and analysis.
Intelligent Bio-Systems president and CEO Steven Gordon, Ph.D., will give a presentation at the upcoming “XGEN” conference. Dr. Gordon will describe a sequencing system that can produce diagnostic-quality test results on multiple samples without the need for batching or bar-coding.
Intelligent Bio-Systems’ three-step sequencing-by-synthesis technology involves amplifying DNA fragments, attaching them to a DNA sequence primer, and then immobilizing them in a high-density array on a glass chip. Fluorescently labeled bases (a different color for A, C, G, and T) are then introduced and attach to the growing DNA strand. The array is scanned and the fluorescent signal emitted by each replicating strand indicates with base was incorporated at the completion of each base addition step.

Complete Genomics reports that its commercial genome sequencing center provides high-quality and low-cost DNA sequencing. The technology consists of three components: DNA nanoarrays, assembled into flow sides; a liquid-handling robot; and a high-speed imager.







