November 15, 2009 (Vol. 29, No. 20)
Heather Holemon Ph.D.
Victoria Rusakova
Use of the TRC Library with Lentiviral Knowledge and Expertise Helps Lead to Successful Results
RNA interference is an effective mechanism for gene silencing whereby double-stranded RNA triggers the cleavage and subsequent degradation of homologous transcript sequences. In this evolutionarily conserved process, longer double-stranded RNA molecules are processed into shorter sequences (21–23 nucleotide small interfering RNAs, or siRNAs) that can bind to the multicomponent RNA-induced silencing complex (RISC).
Within this complex, the siRNA is unwound, and the sense strand is cleaved and dissociated. The antisense strand then remains bound and acts as a guide to target activated RISC to complementary mRNA for cleavage and degradation.
This potent, sequence-specific RNA degradation mechanism was first discovered in plants, where it was termed post-transcriptional gene silencing. It has since been demonstrated in a wide variety of eukaryotic organisms, ranging from fission yeast to humans. These discoveries and subsequent studies into how the RNAi process works have enabled researchers to exploit this pathway and develop tools in order to better elucidate gene function.
Plasmid-based expression of gene-specific small hairpin RNAs (shRNA) under the control of RNA polymerase III–dependent promoters is an effective way to trigger this process. With this approach, the shRNAs are processed intracellularly by the enzyme Dicer into siRNAs, which are then able to directly engage RISC.
The Mission TRC1 shRNA libraries from Sigma-Aldrich consist of over 150,000 such plasmid-based shRNA constructs targeting 15,000+ human and 15,000+ mouse genes. The shRNA sequences are designed using an algorithm developed by the Broad Institute of MIT and Harvard.
On average, there are five shRNA designs for each gene target. The shRNA plasmids are further processed into lentiviral particles to facilitate stable gene silencing in both dividing and quiescent cells. The LentiPlex libraries were generated from the RNAi Consortium’s TRC1 library and are intended for rapid, whole-genome, pooled RNAi screening projects.
While individual genes can be efficiently and robustly targeted using arrayed lentiviral libraries, pooled shRNA libraries may be used to rapidly conduct many phenotypic screens. These pooled screens are typically set up such that the majority of cells have been transduced with a single shRNA to aid in downstream deconvolution.
Positive selection screens are an example of the type of screen that may be conducted using pooled libraries. Selection of a desired phenotype is the basis of this type of screen where cell viability/survival is a commonly used phenotype. After selection, integrated shRNAs are identified by traditional Sanger or deep sequencing methods. The identity of potential hits can then be used to develop hypotheses regarding the biological role of the corresponding gene(s). As with all screens, validation of leads by independent methods such as siRNA, small molecule inhibition, or gene knockout will be required.
The Mission LentiPlex Human Pooled shRNA Library and the Mission LentiPlex Mouse Pooled shRNA Library are genome-wide lentiviral pools. Representation of individual shRNAs from each library is tested before product release to ensure robust library coverage. The actual number of clones in each subpool may vary. Each library is provided as 2 x ~25 µL aliquots of ready-to-use lentiviral format at titers of at least 5 x 108 TU/mL via p24 assay and is predivided into ten subpools of approximately 8,000 shRNA constructs each. Amplification and sequencing primers are also provided for downstream hit identification.
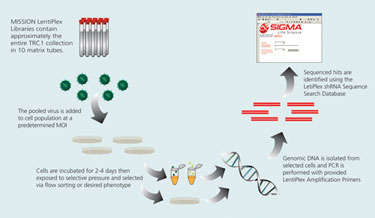
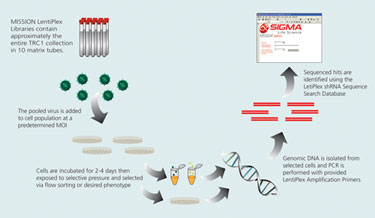
Overview of Pooled Library Screen
There are five primary steps required for utilizing the LentiPlex-pooled library. The following is a brief description of each step.
1. Optimization of puromycin selection conditions: To effectively eliminate colonies with no shRNA insert, optimization of puromycin selection conditions is suggested for each new cell line tested. This can be accomplished by performing an antibiotic kill curve to determine the optimal concentration of puromycin needed to eliminate untransduced cells.
The lowest concentration of puromycin that kills all untransduced cells should be utilized for subsequent experiments. Utilizing higher concentrations of puromycin may lead to unacceptably high cytotoxicity and potential off-target effects.
2. Optimization of transduction efficiency: To successfully identify shRNA sequences of interest, it is critical to set up the screen such that each cell receives only one shRNA construct. By using a low multiplicity of infection (MOI), the probability of multiple integrants per cell is greatly decreased. However, transduction efficiencies and, therefore, desired MOIs depend strongly on the target cell type.
Therefore, it is imperative that determination of the optimal MOI is carried out before starting the screen in a new cell type.
The optimal MOI can be determined by testing a range of MOIs using either Mission pLKO.1-Puro or Mission TurboGFP Control Transduction Particles on a fixed cell density.
3. Transduction and selection: After the optimal MOI is determined, cells are transduced with the viral pools. Transduced cells may be selected using puromycin before the screen is initiated depending upon the type of screen being performed. For screens utilizing a reporter enzyme, the puromycin selection step will eliminate untransduced cells, minimizing the number of cells that will need to be sorted in the end, thereby, maximizing the dynamic range of the assay. For screens assessing changes in viability, the puromycin selection step may not be required if the selective pressure is sufficient to eliminate untransduced cells.
Another variable is the length of time between transduction/
puromycin selection and application of selective pressure. Shorter times will result in fewer cells to maintain and sort at the end. Longer times will result in a broader more representative screen. In addition, the increased number of replication cycles will detect more genes that influence pathways downstream of their immediate targets. This balance will be screen-specific and depend on the type of targets that are desired at the end of the screen. It’s always important to include the most appropriate positive and negative controls for your screen.
4. PCR amplification: Total genomic DNA is isolated from the selected and expanded cell populations. The shRNA inserts can then be readily amplified using the primers provided. The provided Mission shRNA Human Positive Control Vector should be used as template in a separate reaction in order to ensure that the PCR reactions are working optimally.
5. Identification of positive hits: Sequence analysis of the PCR amplicons recovered from cells expressing the phenotype of interest can identify hits from the positive selection screen. The sequencing primer is provided in the kit.
Mission LentiPlex pooled shRNA libraries combine the RNAi Consortium’s shRNA collection with Sigma’s lentiviral manufacturing expertise to enable genome-wide RNAi screens on your benchtop. The LentiPlex pooled screening system provides enhanced delivery and long-term gene silencing in nondividing and primary cell lines. Rapid, whole-genome RNAi screens are now accessible to any researcher with minimal reagent, time, or capital equipment investment.
Overview of pooled library screen
Heather Holemon, Ph.D. ([email protected]), is research and development manager, and Victoria Rusakova is senior research and development scientist, both in research biotech at Sigma-Aldrich. Web: www.sigmaaldrich.com.