May 1, 2016 (Vol. 36, No. 9)
Kate Marusina Ph.D.
Where Trackless Terrain Once Challenged Biomarker Development, Clearer Paths Are Emerging
In time, the narrow, tortuous paths followed by pioneers become wider and straighter, whether the pioneers are looking to settle new land or bring new biomarkers to the clinic.
In the case of biomarkers, we’re still at the stage where pioneers need to consult guides and outfitters or, in modern parlance, consultants and technology providers. These hardy souls tend to congregate at events like the Biomarker Conference, which was held recently in San Diego.
At this event, biomarker experts discussed ways to avoid unfortunate detours on the trail from discovery and development to clinical application and regulatory approval. Of particular interest were topics such as the identification of accurate biomarkers, the explication of disease mechanisms, the stratification of patient groups, and the development of standard protocols and assay platforms. In each of these areas, presenters reported progress.
Another crucial subject is the integration of techniques such as next-generation sequencing (NGS). This particular technique has been instrumental in advancing clinical cancer genomics and continues to be the most feasible way of simultaneously interrogating multiple genes for driver mutations.
Enriching nucleic acid libraries for target genes of interest prior to NGS greatly enhances the sensitivity of
detecting mutations, as the enriched regions are sequenced multiple times. This is particularly useful when analyzing clinical samples, which generate low amounts of poor-quality nucleic acids.
However, NGS has been limited in its ability to identify gene fusions and translocations, which underlie oncogenesis in a variety of cancers. “These challenges are largely related to the enrichment chemistry used to produce sequencing libraries,” commented Joshua Stahl, chief scientific officer and general manager, ArcherDX.
Most target-enrichment strategies require prior knowledge of both ends of the target region to be sequenced. Therefore, only gene fusions with known partners can be amplified for downstream NGS assays.
Archer’s Anchored Multiplex PCR (AMP™) technology overcomes this limitation, as it can enrich for novel fusions, while only requiring knowledge of one end of the fusion pair. At the heart of the AMP chemistry are unique Molecular Barcode (MBC) adapters, ligated to the 5′ ends of DNA fragments prior to amplification. The MBCs contain universal primer binding sites for PCR and a molecular barcode for identifying unique molecules. When combined with 3′ gene-specific primers, MBCs enable amplification of target regions with unknown 5′ ends.
“AMP is ideal for identifying gene fusions and other driver mutations from FFPE samples,” asserted Mr. Stahl. “Its robust utility was demonstrated for detection of gene fusions, point mutations, insertions, deletions, and copy number changes from low amounts of clinical formalin-fixed, paraffin-embedded (FFPE) RNA and DNA samples.
“Tagging each molecule of input nucleic acid with a unique molecular barcode allows for de-duplication, error correction, and quantitative analysis, resulting in high sequencing consensus. With its low error rate and low limits of detection, AMP is revolutionizing the field of cancer genomics.”
In a proof-of-concept study, a single-tube 23-plex panel was designed to amplify the kinase domains of ALK, RET, ROS1, and MUSK genes by AMP. This enrichment strategy enabled identification of gene fusions with multiple partners and alternative splicing events in lung cancer, thyroid cancer, and glioblastoma specimens by NGS.
Ignyta, a precision medicine company, adopted Archer’s AMP technology in Trailblaze Pharos™, a multiplex assay employed in their STARTRK-2 trial for identifying actionable NTRK, ROS1, and ALK gene rearrangements in solid tumors that can be treated with the novel kinase inhibitor, entrectinib. “Gene fusions are incredibly important in personalized medicine right now,” stated Mr. Stahl. “Archer’s FusionPlex assays are quickly becoming the new gold standard.”
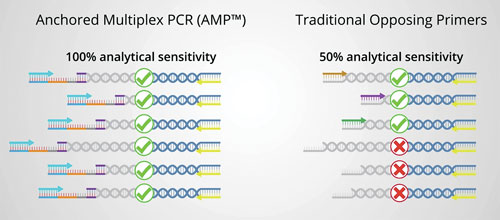
Fusion detection can be carried out with traditional opposing primer-based library preparation methods, which require target- and fusion-specific primers that define the region to be sequenced. With these methods, primers are needed that flank the target region and the fusion partner, so only known fusions can be detected. An alternative method, ArcherDX’ Anchored Multiplex PCR (AMP), can be used to detect the target of interest, plus any known and unknown fusion partners. This is because AMP uses target-specific unidirectional primers, along with reverse primers, that hybridize to the sequencing adapter that is ligated to each fragment prior to amplification.
Reading Cancer Signatures
“Tumor biomarkers are critical for predicting and following patient responses to today’s cancer therapies,” said Darrell Borger, Ph.D., co-director of the Translational Research Laboratory and director of the Biomarker Laboratory, Massachussetts General Hospital (MGH) Cancer Center, Harvard Medical School. “If we understand what drives the malignancy in any given patient, we are able to match existing therapies to the patient’s genotype.”
Over the last decade, the Biomarker/Translational Research Laboratory has focused on developing clinical genotyping and fluorescent in situ hybridization (FISH) assays for rapid personalized genomic testing.
“Initially, we analyzed the most prevalent hotspot mutations, about 160 in 25 cancer genes,” continued Dr. Borger. “However, this approach revealed mutations in only half of our patients. With the advent of NGS, we are able to sequence 190 exons in 39 cancer genes and obtain significantly richer genetic fingerprints, finding genetic aberrations in 92% of our cancer patients.”
Using multiplexed approaches, Dr. Borger’s team within the larger Center for Integrated Diagnostics (CID) program at MGH has established high-throughput genotyping service as an important component of routine care. While only a few susceptible molecular alterations may currently have a corresponding drug, the NGS-driven analysis may supply new information for inclusion of patients into ongoing clinical trials, or bank the result for future research and development.
“A significant impediment to discovery of clinically relevant genomic signatures is our current inability to interconnect the data,” explained Dr. Borger. “On the local level, we are striving to compile the data from clinical observations, including responses to therapy and genotyping. Globally, it is imperative that comprehensive public databases become available to the research community.”
Tumor profiling at MGH have already yielded significant discoveries. Dr. Borger’s lab, in collaboration with oncologists at the MGH Cancer Center, found significant correlations between mutations in the genes encoding the metabolic enzymes isocitrate dehydrogenase (IDH1 and IDH2) and certain types of cancers, such as cholangiocarcinoma and acute myelogenous leukemia (AML).
Historically, cancer signatures largely focus on signaling proteins. Discovery of a correlative metabolic enzyme offered a promise of diagnostics based on metabolic byproducts that may be easily identified in blood. Indeed, the metabolite 2-hydroxyglutarate accumulates to high levels in the tissues of patients carrying IDH1 and IDH2 mutations. They have reported that circulating 2-hydroxyglutarate as measured in the blood correlates with tumor burden, and could serve as an important surrogate marker of treatment response.
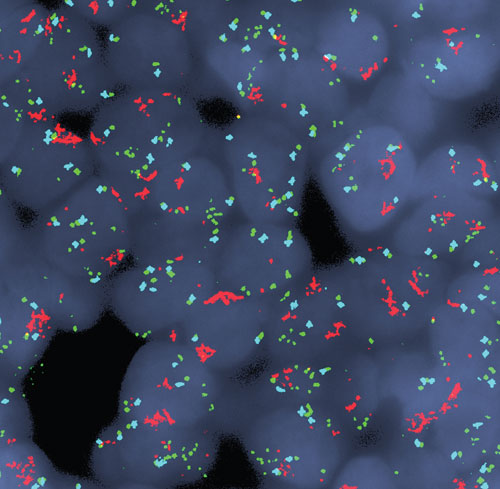
This image, from the Massachusetts General Hospital Cancer Center, shows multicolor fluorescence in situ hybridization (FISH) analysis of cells from a patient with esophagogastric cancer. Remarkably, the FISH analysis revealed that co-amplification of the MET gene (red signal) and the EGFR gene (green signal) existed simultaneously in the same tumor cells. A chromosome 7 control probe is shown in blue.
Tuning Immunosuppression, Preventing Chronic Rejection
“Each year 23,000 kidneys are transplanted, and over 175,000 kidney transplants are functional today,” noted Daniel R. Salomon, M.D., medical program director, Scripps Center for Organ Transplantation, Scripps Research Institute. “However, in just 5 years, 3 out of every 10 patients will be back on dialysis, and in 15 years, at least 75% of all patients will lose their kidney grafts.
“We believe that this is caused by chronic immune-mediated rejection. Failure of effective immunosuppression reduces functional life of these patients and adds in $9–13 billion in yearly healthcare costs.” Dr. Salomon emphasized that ineffective use of immunosuppressive drugs is partially due to the lack of an objective biomarker which could provide decision support for just-in-time adjustment in therapeutic regimens.
“Our research aims to provide that objective measure to clinicians,” explained Dr. Salomon.
To date, kidney transplant biopsies remain the gold standard, even though they are not suitable for continuous monitoring and have both costs and risks. Dr. Salomon’s team developed a minimally invasive diagnostic approach based on unbiased whole-genome expression profiling of blood samples. Using Affymetrix Human Genome U133 Plus 2.0 Gene Chips, the team analyzed 275 blood samples of kidney transplant patients with biopsy-proved acute rejection, acute dysfunction without rejection and transplant excellent phenotype.
The data was passed through several machine-learning algorithms to identify a group of about 250 classifiers that predict subacute or acute rejection with 80% accuracy. This signature is locked while the team continues to expand the core dataset aiming to reach a thousand samples by the end of this year.
“As opposed to classical approaches to biomarker discoveries limited to just a few classifiers, our methodology provides for the first use of unbiased whole-genome profiling in the identification of multiple molecular predictors,” declared Dr. Salomon. “We can use this molecular diagnostic strategy to reveal a subacute rejection prior to significant tissue injury leading to transplant dysfunction. Continuous monitoring would inform physicians on the balance between over-suppression and effective/optimal therapy.”
Dr. Salomon is a chief scientific advisor for Transplant Genomics (TGI), a start-up company created to translate the blood-based molecular diagnostics into clinical tests. In late 2016, TGI will begin providing its TruGraf blood tests for kidney transplant recipients for use by four to six U.S. transplant centers through an early-access program (EAP).
Additional tests designed to be used serially to diagnose and treat subclinical episodes of rejection including biopsy gene profiling are in the final stages of development. Validation and will be made available through the EAP in the upcoming months.
Focusing on Large Molecules
BioAgilytix, a specialized bioanalytical laboratory, is a global leader in large molecule bioanalysis. The company’s business encompasses pharmacokinetic/pharmacodynamic (PK/PD) studies of large biomolecules, in addition to immunogenicity, biomarkers, and cell-based assays. In less than 10 years, BioAgilytix has grown from a start-up to an international powerhouse with over 100 employees—more than half possessing advanced scientific degrees—because of its team’s expertise in the complexities of large molecule drug development.
“In contrast to small molecule analysis, which has become more of a commodity due to its semiautomated and process-oriented nature, large molecule analysis is inherently challenging,” said Afshin Safavi, Ph.D., founder and chief science officer of BioAgilytix. “In large molecule bioanalysis, we rely heavily on analytical reagents, such as antibodies and recombinant proteins, which are known to show considerable variability from lot to lot.
“Therefore, designing an effective analytical process for large biomolecules requires scientific personnel with years of experience. It also requires careful management of critical reagents, and a deep understanding of the capabilities and limitations of the platforms selected for use.”
Dr. Safavi explains that the biomarker field has been trending away from a gunshot approach traditionally favored by large pharma to more focused analyses of a few key biomarkers.
“Unlike several years ago, most biotech and pharma companies now perform careful due diligence and literature research before approaching us, to narrow down their investigation to just a handful of biomarkers,” he explained. Limited samples may drive the desire to multiplex as many biomarkers as possible, but a multiplex approach may often result in low quality data due to reagent cross-reactivity.
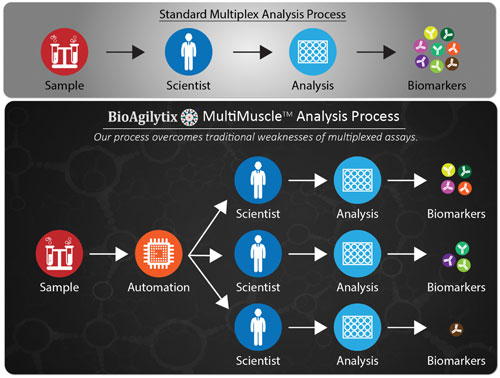
A recent process innovation developed by BioAgilytix, called MultiMuscle Analysis™, uses a customized parallel process to drastically reduce analytical process time and increase data quality. MultiMuscle Analysis splits the sample analysis into multiple parallel tracks, each performed on specialized equipment by scientists experienced in that particular platform.
“Say, for example, a customer requests measurements of 10 biomarkers,” ventured Dr. Safavi. “If we know some of the antibodies may cross-react, then we may, for example, end up with one heptaplex and three as uniplexes, all done in parallel.”
Using this approach, BioAgilytix is able to perform large biomarker analyses on a very large number of samples in near real-time. “We now receive samples from over 20 countries,” Dr. Safavi stated. “We have used the MultiMuscle approach successfully over and over.”
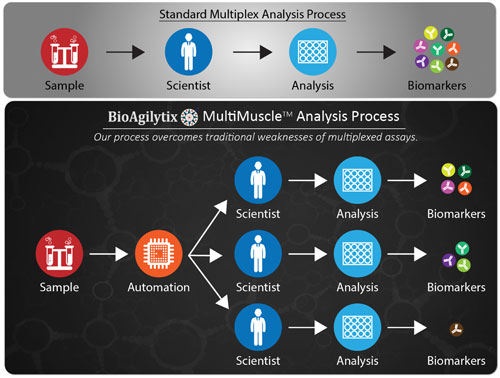
BioAgilytix’ MultiMuscle Analysis is a process that can split sample analysis into multiple parallel tracks to minimize antibody cross-reactivity and allow for use of the best-fit platform or kit for each biomarker analysis. The process may require only one tube of sample with only one F/T cycle.
Predicting Clotting or Hemorrhaging
Venous thromboembolism (VTE) is a disease that includes both deep vein thrombosis (DVT) and pulmonary embolism (PE). It is a common, lethal disorder, symptoms of which are often overlooked. VTE is the third most common cardiovascular illness after acute coronary syndrome and stroke.
Venous thrombi, composed predominately of red blood cells bound together by fibrin, form in sites of vessel damage and areas of stagnant blood flow. Once VTE is diagnosed, anticoagulation therapy is indicated.
A novel anticoagulant that reversibly and directly inhibits factor Xa, a key factor in the coagulation system, has been developed by Daiichi Sankyo. “Once on the path of development of an anticoagulant, we recognized the lack of a rapid and sensitive coagulation test that would not be affected by blood traces of anticoagulant therapies,” said Michele Mercuri, M.D., Ph.D., the company’s senior vice president. “An improved diagnostic test would speed up recognition and treatment of thrombosis, and would aid in development of reversing agents that reduce the effect of anticoagulant therapies when needed.”
When Daiichi Sankyo entered in collaboration with Perosphere to develop a novel broad-spectrum reversing agent, the company also supported development of a point-of-care coagulometer (still under development), a hand-held device designed for broad-spectrum monitoring of the activity of anticoagulants and their corresponding reversing agents, across drug classes. A single test requires only 10 µL of fresh or citrated whole blood from a venous draw or finger stick. It optically measures clotting starting with Factor XII activation to fibrin assembly.
Dr. Mercuri explains that none of the existing tests are able to predict whether a patient is at risk for either clotting or hemorrhaging. “Together with Prof. Zahi Fayad’s Team from the Icahn School of Medicine at Mt Sinai, we used magnetic resonance imaging with the gadolinium-based contrast reagent to detect the venous thrombi and follow their dissolution with edoxaban treatment,” reported Dr. Mercuri.
This study, the edoxaban Thrombus Reduction Imaging Study (eTRIS), was focused on developing and validating a magnetic resonance venography (MRV) image acquisition and analysis protocol for the quantification of thrombus volume in deep vein thrombosis. The multicenter study demonstrated excellent reproducibility of analysis of quantifying thrombus volume.
Sequence and Epigenetic Factors Determine Overall DNA Structure
Researchers at Ulsan National Institute of Science and Technology (UNIST) in South Korea found that DNA molecules directly interact with one another in ways that are dependent on the sequence of the DNA and epigenetic factors.
The researchers found evidence for sequence-dependent attractive interactions between double-stranded DNA molecules that neither involve intermolecular strand exchange nor are mediated by DNA-binding proteins.
“DNA molecules tend to repel each other in water, but in the presence of special types of cations, they can attract each other just like nuclei pulling each other by sharing electrons in between,” explained lead study author Hajin Kim, Ph.D., assistant professor of biophysics at UNIST. “Our study suggests that the attractive force strongly depends on the nucleic acid sequence and also the epigenetic modifications.”
The investigators used atomic-level simulations to measure forces between double-stranded DNA helices, proposing that the distribution of methyl groups on DNA were the key to regulating this sequence-dependent attraction.
The findings from this study were published recently in Nature Communications through an article entitled “Direct evidence for sequence-dependent attraction between double-stranded DNA controlled by methylation.”
The researchers surmised that direct DNA-DNA interactions could play a central role in how chromosomes are organized and packaged, determining the ultimate fate of many cell types.
Dr. Kim concluded by stating that “in our lab, we try to unravel the mysteries within human cells based on the principles of physics and the mechanisms of biology—seeking for ways to prevent chronic illnesses and diseases associated with aging.”