November 15, 2010 (Vol. 30, No. 20)
Patricia C. Stepp, Ph.D.
Kirk A. Ranno
Matthew M. Ferris, Ph.D.
Flow Cytometer Designed to Improve Measurement in Liquid Samples
Virus quantification is of great importance for commercial and academic laboratories involved in research or production of viral vaccines, recombinant proteins, viral antigens, or antiviral agents. The most commonly used methods for virus quantification include the plaque titer assay, 50% tissue culture infectious dose (TCID50), fluorescent focus assay (FFA), and transmission electron microscopy (TEM).
While more modern methods, including quantitative real-time PCR (qPCR) and enzyme-linked immunosorbent assays (ELISA) are becoming more frequently used, there remain significant drawbacks associated with all of these assays.
For example, plaque titer assays require between 4 to 10 days to provide a measure of infectious counts. Likewise, while qPCR may be able to provide results within a single day, it is laborious, requires a skilled operator, and is sensitive to contamination. In general, there is still a need for new analytical methods that can rapidly quantify viral concentration to reduce costs and alleviate bottlenecks associated with current assays.
InDevR has developed a personalized flow cytometer, the Virus Counter®, for rapid virus quantification in liquid samples. This instrument and assay represent a time and cost-effective alternative for the determination of virus concentrations. The assay uses two dyes—one specific for proteins and one specific for nucleic acids—to stain virus samples.
Samples prepared with these universal reagents are quantified by the Virus Counter instrument, which measures the concentration of virus particles (vp/mL) based on the number of particles producing simultaneous events on each of the two distinct fluorescence channels and the measured sample flow rate.
Virus Counter analysis requires ~10 minutes per sample. Figure 1 is a screenshot from the Virus Counter’s InCyt software showing fluorescence signals for the protein (red) and nucleic acid (blue) channels for an influenza A/California/4/2009 (H1N1) sample. The larger upper window shows a 3 ms segment of data and the smaller rectangle box in the lower portion of Figure 1 shows 75 ms of elapsed data. The upper display includes the nucleic acid and protein threshold values (indicated by the overlapping horizontal lines) used to discriminate virus events from background noise.
The Virus Counter has been used to quantify a wide range of viruses, matrix purity levels, and virus concentrations. Some of the most commonly analyzed viruses include adenovirus, baculovirus, coronavirus, cytomegalovirus (CMV), dengue virus, herpes simplex virus (HSV), influenza A (H1N1), influenza B (Flu B), parainfluenza, respiratory syncytial virus (RSV), and rubella.
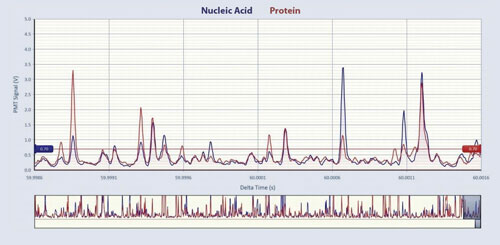
Figure 1. A partial screen capture from the Virus Counter’s InCyt software showing raw data for an influenza A/California/4/2009 (H1N1) sample dilution (~4 x 108 vp/mL). The raw PMT signals for the nucleic acid (blue) and protein (red) channels can be seen in both the large window that displays 3 ms of data and the small window at the bottom, which represents 75 ms of data.
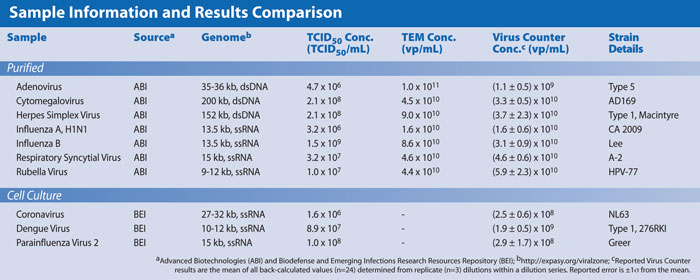
In this study we choose several of these most common virus applications for which commercial sources of well-characterized viruses were available to demonstrate the relationship between Virus Counter results with those from other assays. Sample details, including suppliers and information provided with each sample, are provided in the Table.
High-concentration virus stocks were obtained from either Advanced Biotechnologies (ABI) or Biodefense and Emerging Infections Research Resources Repository (BEI). Samples purchased from ABI (adenovirus, CMV, HSV, H1N1, Flu B, RSV, and rubella) were all purified by the manufacturer with both TCID50 and TEM results provided. Samples from BEI (coronavirus, dengue, and parainfluenza) were unpurified cell culture samples with only TCID50 results provided. Virus genomes ranged from 10–200 kilobases (kb) and represent a broad range of single- and double-stranded RNA and DNA viruses.
In order to evaluate the accuracy of the Virus Counter measurement, a dilution series (5–10,000x) for each virus stock was prepared and each dilution was quantified using the Virus Counter. Results for each diluted sample were compared with calculated results, determined from the reported stock concentrations and the dilution factor used to prepare each sample.
An example log-log plot, showing the relationship of Virus Counter results to TEM and TCID50 results for H1N1 is shown in Figure 2. This data shows that Virus Counter results correlate well (R2=0.98 and slope=0.93) with both expected TEM and TCID50 results over a broad range of virus concentration. These results were also used to back calculate the stock concentration of H1N1 (1.6 ± 0.6) x 1010 vp/mL, which is in agreement with the manufacturer’s reported TEM titer (1.6 x 1010 vp/mL).
Similar dilution analyses were done for each virus, with all samples yielding linear regression fits to the data with R2≥0.98”, slopes between 0.94 and 1.40, and back calculated Virus Counter results listed in the Table. The R2 values and slopes confirm a strong positive correlation and linear relationship between Virus Counter results and the dilution values calculated from the reported TEM and TCID50 concentrations.
In general, Virus Counter results are greater than TCID50 and similar to or lower than TEM values. For example, RSV with a genome size of 15 kb ssRNA had a Virus Counter concentration of (4.6 ± 0.6) x 1010 vp/mL, which was equal to the TEM result (4.6 x 1010 vp/mL). CMV, with a genome size of 200 kb dsDNA, measured (3.3 ± 0.5) x 1010 vp/mL on the Virus Counter, which was only slightly lower than the TEM concentration of 4.5 x 1010 vp/mL. These results demonstrate the ability of the Virus Counter to quantify virus samples with a broad range of genome diversity in a fraction of the time required by standard assays.
It should be noted that while this study was conducted with purified commercially available samples, the Virus Counter has successfully analyzed virus samples from a variety of sources. These real-world samples include viruses in more complex matrices provided by customers and collaborators.
For example, a baculovirus sample provided by a collaborator in centrifuge-clarified growth media with a reported plaque titer of 1.7 x 108 pfu/mL, measured (5 ± 4) x 109 vp/mL on the Virus Counter and provided a dilution series with excellent correlation and linear relationship (R2=1.0 and slope=1.23) to the plaque titer assay results.
In summary, the Virus Counter and associated assay represent a newly available tool for rapid and cost-effective quantification of viruses in liquid samples. The Virus Counter provides a quantitative measure of the number of virus particles per unit volume (vp/mL). The staining process is not virus specific and can be used for the analysis of viruses with highly variable sizes and morphology. Furthermore, the assay, instrument and software are straightforward and do not require a highly trained user.
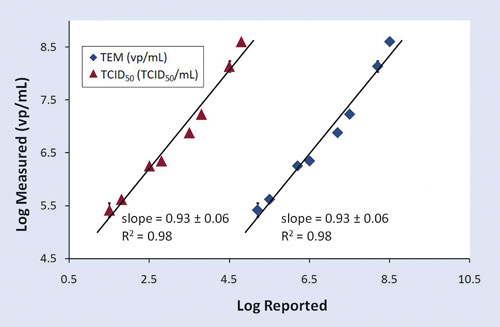
Figure 2. Comparison of log-scale assay results for eight dilutions of influenza A/California/04/2009 (H1N1). Virus Counter results for H1N1 versus TCID50 concentration are shown with red triangles, and H1N1 versus TEM concentration results are shown with blue diamonds. Reported Virus Counter results are mean values (n=3) with the error bars indicating ±1σ from the mean.
Patricia C. Stepp, Ph.D. ([email protected]), is application scientist, Kirk A. Ranno is research assistant, and Matthew M. Ferris, Ph.D., is lead scientist, instrument development, at InDevR.







