February 1, 2010 (Vol. 30, No. 3)
Todd Watterson
Multiplex Systems that Conserve Time, Money, and Samples Are Now Available
The protein-detection landscape has seen a number of advances in the last decade. Much of our knowledge of proteins has come from using ELISA, Western immunoblot, immunoprecipitation, and two-hybrid systems. Of those advances, the ability to simultaneously detect multiple proteins in a single sample has greatly enhanced our understanding of some of the complex protein interactions that occur in normal and disease states. The manifold benefits of multiplex protein detection are increasingly being appreciated by researchers.
Validation of genomic data is a major area where rapid multiplex protein detection is needed. Many researchers are now familiar with instances where alterations in mRNA do not equate to changes in protein levels. Some protein levels are increased by mRNA stability rather than increased mRNA transcription.
There are examples where a treatment or disease state triggers endoplasmic reticulum stress, which results in the blockage of translation of many proteins that are not related to protein folding, independent of mRNA upregulation. Others include instances where differential splicing and post-translational modifications are required for protein activity rather than mere protein presence, such as phosphorylation of Akt, or cleavage of interleukin-1 and tumor necrosis factor from inactive to active proteins. Proteomic mapping and biomarker discovery are other areas enhanced by multiplex protein detection.
Researchers’ demand for the obvious advantages of multiplex protein detection has fostered the entrance of multiple technologies and platforms into the research marketplace. Protein-detection technologies can be differentiated by the direct use of antibodies (e.g., ELISA, reverse-phase arrays) and those that do not use antibodies (e.g., mass spectrometry).
The systems that utilize antibodies are less expensive, and therefore, have obtained greater market adoption. Most antibody-utilizing technologies are derivatives of ELISA. Many are sophisticated technologies, however the high cost and need for high-maintenance equipment has limited their use. Fortunately, there are lower cost solutions that offer as good, if not better, quality in multiplex protein detection.
Quansys Biosciences has developed a multiplex protein array technology, Q-Plex™ multiplex ELISA, that enables high-quality low-cost multiplex protein detection. Using modern liquid-dispensing systems that are capable of precisely dispensing nanoliter volumes of liquid, multiple proteins are spotted down into the well of standard 96-well plates.
For sandwich-formatted ELISAs, the proteins spotted down are capture antibodies. The capture antibodies are separated from each other by their location on a Cartesian coordinate system allowing multiple ELISAs (multiplex ELISA) to be performed in a single well. The sandwich ELISA is performed with two cocktails, an antigen standard and biotinylated secondary or detection antibodies. A luminescent signal generated by infrared (IR) fluorescence or chemiluminescence is imaged by either an IR scanner or CMOS or CCD camera with the luminous intensity being proportional to the amount of antigen bound by the antibodies.
The use of chemiluminescent or IR fluorescent based detection systems in the Q-Plex ELISA enables most laboratories to use readily accessible multifunctional equipment negating the need to purchase additional expensive single-function equipment. The images are then analyzed with software to quantify the pixel values and perform regression analysis giving actual quantitative values for the unknown samples.
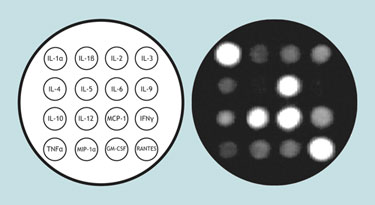
Pattern of cytokine release in mice following influenza infection
Advantages of the System
The advantages of the Q-Plex ELISA include time, sample, and cost savings. Multiplex ELISA is an evolutionary step from a traditional ELISA involving similar steps of antigen/sample incubation, secondary or detection antibody incubation, IR fluorescent or chemiluminescent label incubation, wash steps, and plate reading resulting in a 2.5 hour assay time.
Nonsandwich ELISAs such as competitive (a single capture antibody is placed on the plate and a labeled antigen is used that competes with the antigen in the sample for binding with the antibody) or direct ELISAs (the antigen is on the plate rather than a capture antibody) have been designed and multiplexed that require as little as 30 minutes total assay time.
When the time it takes to run a single multiplex ELISA is compared to the time required by multiple standard ELISAs, the time savings is apparent. Similar logic applies to sample volumes. Running 16 simultaneous ELISAs in a single well (the sample amount required for a Q-Plex ELISA is 30 µL per well) greatly reduces the amount of sample required to obtain the same results.
While multiplexing technologies for protein detection are useful for screening which proteins are in a specific sample, a less obvious but potentially more significant use is for the establishment of protein-expression patterns. Such patterns might eventually lead to the discovery of better biomarkers, especially in circumstances where a single suitable biomarker has yet to be found.
For example, cytokines are generally low-level transitory signaling molecules that are tightly regulated. However, they are differentially upregulated and released depending upon the stimulus. Using a mouse cytokine Q-Plex ELISA measuring 16 different cytokines and chemotactic factors researchers have found that influenza virus stimulates a rather consistent pattern of cytokine release in mice.
The major upregulation of RANTES, interleukins 1α, 6, 12, and monocyte chemotactic protein-1 in mice following influenza virus infection is shown in the Figure, with lesser alterations in the other proteins detectable by the array. While this is far from sufficient for diagnostic purposes it does demonstrate that protein-expression patterns are elicited from a virus, and that even patterns of low-abundance transitory proteins are detectable.
Other Uses
Other uses of multiplex protein-detection technologies include the reverse-phase array. Analogous to miniaturized dot blots, homogenized protein is arrayed and probed with antibodies. With reverse-phase arrays the protein can be spotted at differing concentrations and EC50 values can be calculated to determine antibody binding efficiency.
Alternatively equal concentrations of different protein homogenates can be spotted and probed with antibodies to specific proteins, for example, tissue homogenates from cancer biopsies and healthy tissue or cell lysates from treated and untreated cells can be arrayed next to each other and examined for relative protein expression.
There remain many areas in the still immature multiplex protein-detection market where improvement is warranted. Expanded use of nonlinear regression analysis allows for better modeling of the type of standard curves generated by antibody-based technologies, but not all of the standard curves in an array are best explained by a single model.
As the number of standard curves generated in a protein array increases, the need for automated regression of multiple curves and determination of optimal curve fit becomes more apparent. Also, large amounts of data are generated with protein arrays and much of the sifting, prioritizing, and interpreting of the data remains in the hands of the researcher using many of the same tools that would be used to analyze standard ELISA data.
There are few software options for protein-array analysis that have the sophistication of those used for genomic arrays offering features such as pathway and statistical analysis. These shortcomings will likely be addressed as the multiplex protein-detection market expands and bioinformaticians notice its relatively untapped potential.
As there is no single dominant technological platform for multiplex protein detection, researchers are presented with multiple alternatives. With the establishment and proven functionality of lower cost multiplex technologies, the value of multiplex protein detection will likely be realized by more researchers. Most biological science laboratories will probably adopt multiplex protein detection, whether in their own laboratories or through the use of off-site sample testing services, within the next few years.
Todd Watterson ([email protected]) is technical sales scientist at Quansys Biosciences. Web: www.quansysbio.com.







