February 1, 2010 (Vol. 30, No. 3)
Richard A. A. Stein M.D., Ph.D.
Findings Could Transform Cancer Biology and Provide a Host of Benefits
The biomedical literature provides many examples when medical conditions that were classically viewed as a paradigm for genetic inheritance appear to be shaped by additional factors, which cannot be explained by DNA sequence variation alone. For example, there are several reports of monozygotic twins with Huntington disease who, despite harboring identical numbers of the CAG trinucleotide repeats, differ in their age at onset and in clinical presentation and progression.
In addition to the genetic information required to establish an organism, recent decades have unveiled a previously unknown type of chromatin modification, known as epigenetic, which is defined as heritable DNA changes that are not encoded in the sequence itself. Unlike genetic modifications, the epigenetic ones are reversible, and increasingly appear to serve fundamental roles in cell differentiation and development.
An important group of epigenetic modifiers is the polycomb group of proteins. They were first described in Drosophila melanogaster, are conserved across species and function as critical regulators of several genes from embryogenesis to adulthood. Polycomb proteins reside in two multiprotein assemblies—polycomb repressive complexes PRC1 and PRC2, of which PRC2 is the only one currently known to di- and tri-methylate lysine 27 of histone H3—to establish repression of target genes. For this reason, the PRC2 complex has attracted considerable attention.
“This complex is very exciting from the basic science point of view, it is important for cancer and, in addition, serves as the paradigm to understand the basic regulation of transcription,” says Kristian Helin, Ph.D., professor and director at the Biotech Research and Innovation Centre at the University of Copenhagen.
One research effort in Dr. Helin’s lab focuses on understanding the biochemical organization and the biological function of this complex. Previously, Dr. Helin and collaborators showed that trimethylated histone H3 lysine 27 chromosomal marks that recruit PRC2 complexes are subsequently maintained on daughter strands during DNA replication. The team has proposed a model to explain how this epigenetic mark is maintained during interphase and inherited during cell proliferation, to preserve the gene-expression profile and the cellular identity during subsequent generations.
“One key question is how these epigenetic modifications are targeted and recruited to specific locations, and how they regulate cellular events,” explains Dr. Helin. “In addition, an important question is whether this complex is going to be a good marker for certain types of cancer.”
“Epigenetics in general, not just DNA methylation, is going to be very powerful. The more we are learning about chromatin marks, the better we are able to understand the behavior of tumors and predict their aggressiveness,” explains Stephen B. Baylin, M.D., professor of cancer research and deputy director of The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University.
In most tissues, CpG islands around transcription start sites are largely unmethylated, but their methylation has been described in many tumors and can serve as potential biomarkers. One of the advantages of using epigenomic biomarkers is that, in most cases, DNA methylation changes precede clinical symptoms. “If there is a small abnormality that is not yet an invasive cancer, but a precancerous lesion, or a small tumor, many genes will have abnormal methylation, and that is probably true in many tissues,” says Dr. Baylin.
Recent work conducted by Nita Ahuja, M.D.’s lab, located in Dr. Baylin’s group, in collaboration with investigators from Johns Hopkins Medical Institute, the National Institutes of Health, and Vrije University from Amsterdam, revealed that TFPI2 promoter methylation occurs as an early and frequent event during colorectal cancer, and promises to become a valuable epigenetic marker when used in combination with other noninvasive colorectal carcinoma screening tools.
Another exciting application of epigenetic markers that emerged from a study led by Malcolm Brock, M.D., in the Baylin program, is in the molecular staging of malignant tumors. By examining abnormal methylation patterns in both the tumor and the lymph nodes of patients with non-small-cell lung cancer, a malignancy that frequently recurs subsequent to treatment, Dr. Brock and collaborators reported that promoter methylation of several genes, even in patients with stage I cancer with histologically negative lymph nodes, was associated with tumor recurrence.
In addition, methylation of both p16 and CDH13 in the tumor and mediastinal lymph nodes was associated with a 15-fold higher relative risk of recurrence. “These tumors behaved, not like stage I, but like stage III tumors,” explains Dr. Baylin. “These results need to be validated in larger studies, and this is an example of using molecular biomarkers for the molecular restaging of the cancer, a very powerful approach. Biomarkers based on DNA methylation and other chromatin marks are going to be very useful, in the future, to grade tumors.”
Molecular Model
The ability to therapeutically target epigenetically silenced genes requires a detailed knowledge about the molecular mechanisms of gene repression. Dr. Baylin and colleagues recently presented a molecular model to explain how DNA methylation causes gene silencing in mammalian cells.
The authors used the GATA-4 gene as a model to investigate how polycomb protein complexes and DNA methylation maintain the chromatin in its silent state. They found that polycomb protein occupancy at genomic regions enriched in trimethylated histone H3 lysine 27 marks establishes long-range interactions by chromatin looping. “If the cell expands without maturing properly, the polycomb proteins appear to remain in place, and they are the ones that appear to cause the looping in a way that represses the gene. If DNA methylation is added to that, the loops become tighter, and gene repression is tighter,” explains Dr. Baylin.
This finding promises to significantly improve our understanding of higher order chromatin organization and gene silencing both in stem cells and in cancer cells, which share intriguing similarities with respect to chromatin organization.
Therapeutic Perspectives
One of the PRC2 components, the catalytic protein EZH2, was previously shown to be highly expressed in the most aggressive types of cancer, very much in agreement with the observations that this protein fulfills a central role in carcinogenesis.
In collaboration with investigators from the National Institutes of Health and Novartis Institute for Biomedical Sciences, Kapil N. Bhalla, M.D., professor of medicine and director of the Medical College of Georgia Cancer Center, recently showed that the combined treatment with 3-Deazaneplanocin A, an S-adenosyl-L-homocysteine hydrolase inhibitor, and panobinostat, a pan-histone deacetylase inhibitor, caused more depletion of EZH2, and of trimethylated lysine 27 marks on histone H3, than either compound alone.
In addition, the two compounds synergistically increased apoptosis in cultured and primary acute myeloid leukemia cells, but not in normal bone marrow progenitor cells. “This opens up the very exciting possibility that perhaps combining histone deacetylase and methytransferase inhibitors represents a good therapeutic option to target epigenetic modifications in acute leukemia, and perhaps in lymphomas, too, because EZH2 was also implicated in some lymphomas,” predicts Dr. Bhalla.
In addition to the work on acetylation and methylation in the context of cancer epigenetics, another interest in Dr. Bhalla’s group focuses on the involvement of heat shock proteins (Hsp) as molecular chaperones in cancer. Cancer cell metabolism creates a considerable amount of stress, and one of the main categories, known as proteotoxic stress, is mediated by the misfolded or unfolded protein response. Heat shock proteins are essential in maintaining the correctly folded conformation and activity of oncoproteins, and thus allow cancer cells to survive the stress response.
A few years ago, Susan Lindquist, Ph.D., and collaborators from the Massachusetts Institute of Technology and Howard Hughes Medical Institute, showed that deletion of heat shock factor 1, the master eukaryotic heat shock regulator, protected mice from tumors induced by mutations in the RAS oncogene or the p53 tumor suppressor gene, indicating that heat shock proteins are essential for cancer development.
The use of molecular chaperones as therapeutic targets for malignant tumors emerges as an exciting idea, and several Hsp90 inhibitors are currently being investigated as potential anticancer agents.
In collaboration with Ari Melnick, M.D., from Cornell University, Dr. Bhalla recently showed that Hsp90 inhibitors selectively kill diffuse B-cell lymphomas by targeting the Bcl-6 transcriptional repressor, a common oncoprotein that represses several tumor suppressor genes. The therapeutic potential of Hsp90 inhibitors is especially valuable in context of the high toxicity of certain regimens currently used to treat lymphomas. “With Hsp90 inhibitor, the possibility to modulate the proteotoxic stress phenotype of cancer cells, used in combination with a targeted agent against the oncoprotein kinase, to which the cancer is addicted to, would constitute a very attractive therapeutic option,” says Dr. Bhalla.
Smoking and Infectious Diseases
“Our individual life history is inscribed in our epigenome,” states Toshikazu Ushijima, Ph.D., chief of the carcinogenesis division at the National Cancer Center Research Institute, Tokyo. Dr. Ushijima and collaborators screened genes that were silenced in esophageal squamous cell carcinomas and demonstrated that methylation levels in five promoters are significantly correlated with the duration of tobacco smoking, indicating that chronic smoking induces methylation changes in many of these genes.
This finding supports the idea that smoking induces an epigenetic field of cancerization, a term that was previously described for breast, colon, liver, and stomach cancers, and is used to denote epigenetic modifications that occur during the early stages of carcinogenesis.
Back in 2006, Dr. Ushijima and colleagues used real-time methylation-specific quantitative PCR to conduct a temporal characterization of DNA-methylation levels in the gastric mucosa in patients infected with Helicobacter pylori, a pathogen that represents a major cause of gastric cancer in several countries worldwide.
The investigators revealed that methylation of promoter CpG islands was high in individuals infected with Helicobacter pylori, and that high methylation levels correlated with the subsequent risk to develop gastric cancer. This pointed toward an epigenetic field for cancerization established as a result of the infection and measurable by DNA-methylation levels. Methylation levels decreased after the bacteria were eradicated but were still much higher as compared to individuals without the infection during their lifetime. “The epigenetic field defects are becoming more and more important,” says Dr. Ushijima.
Increasingly, new revelations about epigenetic modifications promise to transform all facets of cancer biology and to provide prophylactic, diagnostic, and therapeutic benefits. Epigenetic modifications could, in addition, become one of the missing links between infectious diseases and cancer.
The ability of certain viruses, bacteria, parasites, and protozoa to cause malignant transformation represents one of the most fascinating topics in life sciences. This connection was regularly re-discovered throughout the past century, it repeatedly fell into oblivion and, historically, demonstrating causality often proved challenging.
It is currently estimated that approximately 20% of all cancers worldwide are linked to pathogens, and the involvement of epigenetic changes in shaping this connection could soon lead to new chapters in cancer biology, establishing links that we never would have thought could exist.
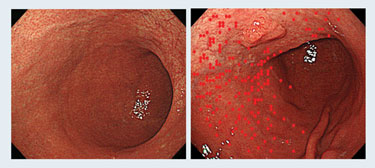
Researchers at the National Cancer Center Research Institute in Tokyo have been studying DNA-methylation levels in the gastric mucosa of patients infected with Helicobacter pylori for several years.
Sidebar: Bisulfite Treatment for the Detection of DNA Methylation
Sodium bisulfite can deaminate or convert cytosine in DNA into uracil, but does not affect 5-methylcytosine. Bisulfite treatment of DNA is a prerequisite for DNA-methylation analysis for many epigenetics-based studies involving methylation profiling and the quantification of methylation status.
However, analytical procedures involving bisulfite-treated DNA are subject to variability due to DNA degradation, incomplete conversion, and low yields of DNA.
A team of scientists from Zymo Research investigated the procedure of bisulfite treatment of DNA paying particular attention to the chemistries involved in the process and to conversion rates to limit variability between samples and improve upon conventional methods.
They report that conventional bisulfite DNA conversion chemistries could be improved without the levels of DNA degradation typically resulting from incubation of reaction mixtures at high temperature and nonphysiological pH. Essential to this process was prohibiting the overconversion of 5-methylcytosine into uracil that can occur in some situations and reaction conditions.
The researchers found that the bisulfite conversion process could be simplified and the variability between treatments kept to a minimum by coupling heat denaturation with the conversion process and by using in-column desulphonation to clean and purify the converted DNA. This new method was found to yield an average of >80% recovery of input DNA with >99% cytosine to uracil conversion.
The method has been specifically designed to accommodate (in addition to purified DNA) biological fluids, cells, or tissue directly as the input material. This makes its application for FFPE and LCM-derived samples particularly well suited, according to the Zymo Research investigators.

Many technologies from Zymo Research are compatible with the workflows of systems/platforms used for DNA methylation analysis and quantitation.
Application Note
In an application note (“Perfecting Bisulfite Treatment for DNA Methylation Detection”), the scientists described the use of the EZ DNA Methylation-Direct™ kit for the recovery of bisulfite-treated DNA from a range of sample inputs. They noted that the kit and associated reagents were designed to achieve data consistency and mitigate the loss of DNA. According to the team, the technologies involved in the kit ensure consistent recovery of input DNA from as few as 10 cells or as little as ~50 pg DNA.
“This is facilitated through the fine-tuning of bisulfite conversion chemistries that enable the reaction to proceed to near completion, that is, 99.8% conversion of nonmethylated cytosine, while maintaining the integrity of methylated cytosine during the process,” says Marc Van Eden, Ph.D., vp business development and marketing, and a co-author of the application note.
Dr. Van Eden maintains that the key to the high recovery of converted DNA is the advanced column/plate design featured in the company’s kit.
“The Fast-Spin columns and plates ensure rapid desulphonation as well as high recovery of converted DNA. Elution of buffers from the column/plate matrices is complete, negating buffer carryover,” continues Dr. Van Eden. “Thus, eluted, bisulfite-converted DNA is pure and ready for analysis. Further, the columns allow DNA to be eluted in ultrasmall volumes (≥6 µL) for highly concentrated DNA if required.”







