September 15, 2015 (Vol. 35, No. 16)
Dara Grantham Wright Senior Vice President and General Manager eBioscience, a business unit of Affymetrix
New Techniques Enable Closer Look into Genetic & Cellular Alterations in Tumor Microenvironment
For years, researchers and physicians have suspected, and have worked to demonstrate, how the immune system affects susceptibility to, defense against, and progression of certain cancers. It is now understood that the immune system has the ability to influence the fate of developing cancers by not only functioning as a tumor promoter that facilitates cellular transformation, promotes tumor growth, and sculpts tumor cell immunogenicity, but also as an extrinsic tumor suppressor that either destroys developing tumors or restrains their expansion.
In the last few decades, drugs, biologicals, and vaccines targeting certain attributes of the immune system, known as immunotherapeutics, have become available, and emerging clinical data suggest that cancer immunotherapy is likely to become a key part of the clinical management of cancer for years to come.
Although immunotherapies represent a major step forward in cancer care, providing in some cases unprecedented response rates, there is still much work to do to discover new druggable targets, find biomarkers to predict response, as well as gain deeper understanding of why some cancer types are incredibly responsive to immunotherapeutic treatments while others are not.
How Immunotherapies Work
In general, immunotherapies direct an individual’s immune system to fight cancer by either stimulating it to attack cancer cells or by introducing manufactured immune system components to augment immune function. Immunotherapy treatments work in different ways. Some boost the body’s immune system in a very general way. Others help train the immune system to attack cancer cells specifically.
On an immuno-oncological level, the genetic and cellular alterations that define a cancer cell provide the immune system with the means to be recruited to the tumor and generate T-cell responses to recognize and eradicate those cells. Elimination of cancer by T cells is only one step in the cancer immunity cycle. T-cell activation is controlled by both stimulatory and inhibitory checkpoints. Tumors use the expression of inhibitory ligands as a mechanism of suppressing cytotoxic T-cell response and inducing an immunosuppressive environment.
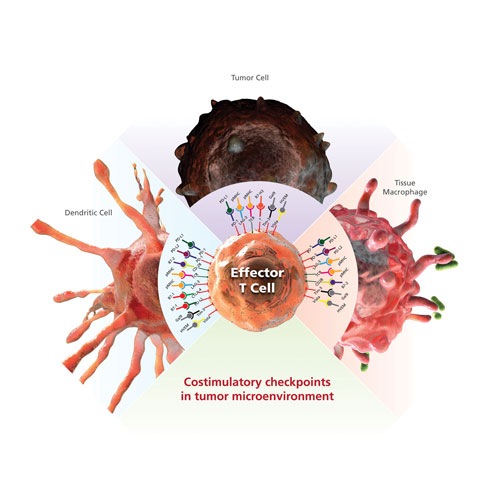
Identification of specific cancer T-cell inhibitory signals, such as PD-L1, has prompted the development of a new class of cancer immunotherapy that specifically hinders immune effector inhibition, reinvigorating and potentially expanding preexisting anticancer immune responses (Figure 1).
The presence of environment-altering immunosuppressive innate myeloid lineages in the tumor microenvironment may further explain the limited activity observed with previous immune-based therapies and why these therapies may be more effective in combination with agents that target other steps of the cycle.
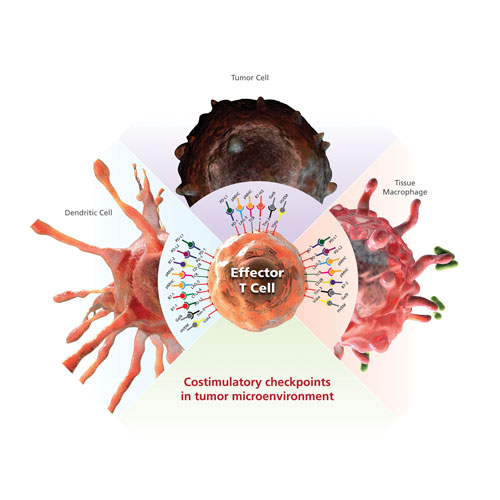
Figure 1. Inhibitory costimulatory checkpoints are a natural immune mechanism for self-tolerance and minimization of collateral tissue damage. Inhibitory checkpoint receptors such as PD-1, LAG-3, TIM-3, and CTLA-4 are expressed by T cells, and their ligands are expressed by macrophage and dendritic cells. Tumor cells can express multiple inhibitory ligands to repress T-cell function and thereby evade clearance by the immune system.
Understanding the Tumor and Its Microenvironment
A deeper understanding of cancer as a disease requires the acknowledgement of its inherent heterogeneity. As with the cancer cells within a tumor, the immunological microenvironments in which they grow are similarly heterogeneous. Emerging and well-established scientific tools and techniques for the analysis of cancer cells, immune cells and their microenvironment can be combined to yield new insights into the nature of tumorigenesis, immune system recruitment, and treatment optimization.
In addition, the presence and quantity of various immune cell types in the tumor microenvironment may have prognostic value. Many scientists believe that a deepening appreciation of oncology genomics and the quantity and type of antigens expressed by the tumor cells, when coupled with an analysis of the patient’s immune system, will greatly progress the field and unlock the next generation of immunotherapies.
Flow cytometry and immunohistochemistry are established tools for the labeling and analysis of immunological and oncology cellular components. New techniques are likewise becoming more widely used that enable simultaneous detection of proteins and nucleic acids at single-cell resolution.
New Cellular Analysis Tools
eBioscience, a business unit of Affymetrix, has recently expanded commercialization of two such novel assays that provide exciting new technologies in the armament of cellular analysis techniques for immuno-oncology research. The first is PrimeFlow™ RNA Assay, which is the only commercially available assay for the simultaneous detection of RNA and protein expression within millions of cells at single-cell resolution using a standard flow cytometer. The assay is compatible with cell surface and intracellular antibody staining, using traditional fluorochromes for multiparameter cellular analysis.
With this technology an immune-oncology researcher could explore gene expression heterogeneity among different rare tumor-infiltrating immune cell subsets with single-cell resolution and without laborious cell sorts, as well as compare kinetics of both RNA and protein in the same cell.
S. Shalapour et al. recently published a study in the journal Nature (April 29, 2015) applying these techniques to mouse models of castrate-resistant prostate cancer demonstrating that the presence of a very specific and rare (0.04–3% of total) B cell population in the tumor microenvironment correlates to a immunotherapeutic response allowing a CTL-dependent eradication of oxaliplatin-treated tumors.
ViewRNA® In Situ Hybridization (ISH) Cell and Tissue Assays comprise the second new technique from eBioscience. Similar to the PrimeFlow RNA assay, but compatible with microscopy, these assays enable the visualization of single-copy RNA transcripts within adherent and suspended single cells or single cells in tissue sections, and in the case of ViewRNA ISH Tissue Assays, the spatial separation of tumor subclones by phenotypic RNA expression. Similarly, this technique can be used to visualize and quantitate cellular and molecular attributes of tumor-infiltrating immune cells to elucidate biomarkers of resistance and response. Leveraging these novel cell analysis approaches, immuno-oncology researchers can analyze cellular diversity in the tumor microenvironment as well as the diversity of immune cell responses at a single-cell level.
Breakthrough responses to new immunotherapies are stimulating a renewed interest in basic immune biology. With our quest to develop strategies to harness the human immune response against cancer to achieve durable responses and/or complete eradication of cancer in patients safely, we must explore multiple approaches simultaneously. Which immune checkpoints can be manipulated? Are there dual therapies that can be applied to improve responses? Are there biomarkers inherent to the immune system in general, the specific tumor and the tumor microenvironment that can be used to stratify responders?
Multiple approaches to cancer therapy exist, and few are as complicated as immune-based therapy. That being said, few therapies in recent history have demonstrated such extraordinary and durable responses for the patients who do respond. As such, many believe that this will be an intensifying area of research and clinical focus for years to come.
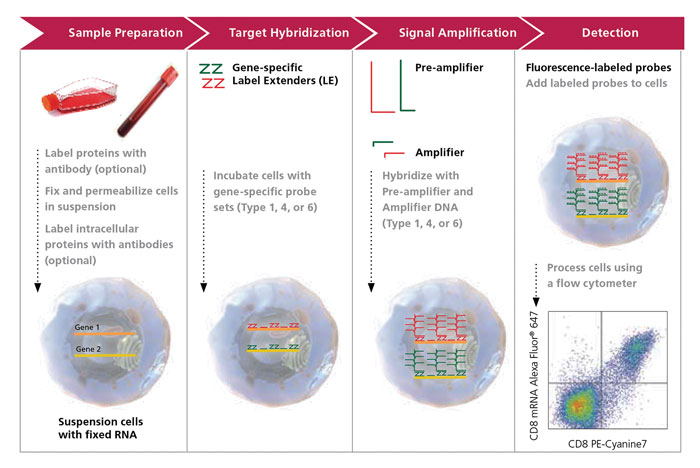
Figure 2. The PrimeFlow RNA Assay workflow contains several steps: antibody staining, fixation and permeabilization including intracellular staining if desired, followed by target hybridization with a target-specific probe set containing 20 to 40 oligonucleotide pairs. Next, branched DNA signal amplification is achieved through a series of sequential hybridization steps consisting of pre-amplifiers, amplifiers, and labeled probes, followed by detection by flow cytometric analysis. This results in excellent specificity, low background, and a high signal-to-noise ratio. For simplicity, two RNA targets are shown in the schematic above (red and green), and only 3 of the 20 to 40 oligonucleotide target probe pairs per target RNA are shown.
Dara Grantham Wright ([email protected]) is senior vice president and general manager, eBioscience, a business unit of Affymetrix.