December 1, 2013 (Vol. 33, No. 21)
Kate Marusina Ph.D.
It seems inevitable that human whole-genome sequencing will dramatically change the practice of healthcare in the next decade. Early disease detection and personalized and directed healthcare interventions are just a few areas where genomic sequencing technology may have a clear value proposition.
But if clinical application of genome sequencing is to become a reality, we still need to reach several milestones. We need to demonstrate that whole-genome sequencing truly has a potential to save both lives and reduce healthcare costs. We need to improve the diagnostic quality and drastically reduce the costs of informatics analysis. We need to develop a regulatory environment similar to what is currently experienced by clinical labs. And we need to enhance the education of all healthcare professionals and generate greater awareness among future users. It feels like a long way to go. Or is it?
A tremendous amount of pilot work is already being done, radically transforming current mechanisms for translating genomic advances into improved healthcare. The contributors to this article describe the pioneering research they are pursuing and how it is bridging genomics and medicine to bring about the rational use of genomics technology in clinical practice.
“Are we ready to use genomic information to change our lifestyles or seek additional medical care?” asks Joseph Jarvis, Ph.D., senior research scientist, Coriell Institute for Medical Research. “The application of the clinical genome is only as valuable as our willingness to use it for healthcare decisions.”
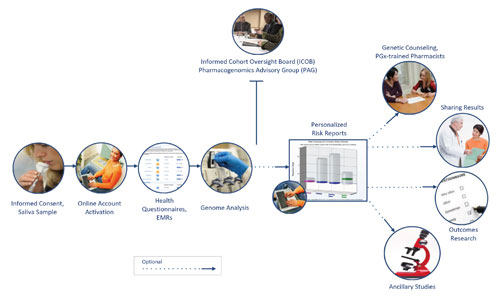
Dr. Jarvis concentrates his efforts on the Coriell Personalized Medicine Collaborative (CPMC), which aims to answer how best to deliver genetic risk information to users.
More than 7,500 individuals are grouped into the CPMC’ s four longitudinal cohorts, including a segment representing the United States Air Force Medical Service, an elite organization eager to learn more about the clinical utility of genetic information. Air Force staff recognized the promise of genome sequencing and engaged Coriell in a train-the-trainer capacity in anticipation of a genome-informed health model.
Participants receive the results of their genome-wide single nucleotide polymorphisms presented in the context of their medical and personal history. “We focus on SNP genotyping,” adds Dr. Jarvis, “because whole-genome data is too difficult to interpret and present to the user in an actionable format.”
Coriell completes genotyping with Affymetrix technology (the Genome-Wide Human SNP Array 6.0 and DMET Plus chips), and it generates reports for presentation to users.
These reports, which detail well-replicated clinical associations, combine genetic and nongenetic risk assessments in an easy-to-read, web-based format. This presentation represents CPMC’s attempt to combine information over which the participants have full control (lifestyle choices) with inherent data (genetics and family history). Enrollees have a choice of whether or not to view reports for a particular disease association.
“One of the study outcomes is to evaluate how the users act on this information,” says Dr. Jarvis. “Opting out from viewing it is also an outcome.” The majority of participants, however, choose to view the results, and some act on information by seeking additional disease-specific testing.
Sharing results with treating physicians is one of the desirable outcomes sought by the CPMC, and the CPMC project uses the opportunity to outline teaching techniques and encourage doctors to incorporate genetic information at the point of care.
A recent Coriell spin-off venture, Coriell Life Sciences, will employ lessons learned from the CPMC project and optimize the doctor-patient exchange by integrating the storage, interpretation, and delivery of genomic information.
Participants in the the Coriell Personalized Medicine Collaborative (CPMC) study provide a saliva sample for DNA analysis in Cornell’s Genotyping and Microarray Center using CLIA-certified genotyping platforms. The CPMC study represents an attempt to broaden informed consent, one facet of translating genomics into better clinical care.
Improving Variant Calling
Rapid advances in DNA sequencing technologies have the potential to inform current and future medical care. To facilitate the broad clinical implementation of next-generation sequencing, it is critically important for genomic medicine to acquire the typical characteristics of clinical diagnostics, including clinical-grade sample collection, high-quality sequencing, and rigorous generation of variant calling.
While the sequencing process itself is by now fairly standardized, the bioinformatic analysis and interpretive schemes are quite variable.
Gholson Lyon, M.D., Ph.D., assistant professor of human genetics at Cold Spring Harbor Laboratory (CSHL), highlighted these differences in the interpretation of sequencing data by comparing five different alignment and variant calling software packages.
Calling of common variants (found in over 1% of population) was relatively consistent. However, software packages called rare mutations with different accuracy. Insertions and deletions (collectively termed “indels”) presented considerable difficulty to all software packages. The position of indels is often ambiguous, and their calling could affect the outcome of clinical diagnosis.
“Indels are an integral part of structural variant analysis,” says Dr. Lyon. “But development of indel-calling tools has been lagging.” To address this issue, Dr. Lyon is working with Michael Schatz, Ph.D., assistant professor of quantitative biology at CSHL, who is developing and validating novel indel mapping strategies.
Dr. Lyon also suggests that de novo identification of family-inherited mutations could be significantly improved by including multiple family members in the sequencing efforts. Sequencing multigenerational pedigrees enables increased accuracy simply because the same region is now read multiple times.
“Millions of genomes should be collated to truly understand what mutations really mean,” says Dr. Lyon. “Collaborative wiki-type annotation of variants will accelerate our understanding of phenotype-genotype relationships. The future of clinical genomics is based on the collective input of the world-wide community of scientists.”
Genome Knowledge Platform
“We have responded to the call for a system to manage the volume and complexity of next-generation sequencing analysis,” says Ben Salisbury, Ph.D., vp of clinical genomics at Knome.
“We have built the knoSYS™ 100, an end-to-end software and hardware platform for developing, running, and sharing in silico genetic tests.”
The knoSYS provides a pipeline that includes sequence alignment, variant calling, interpretation, and reporting. Users can set up their own parameters and filters, and add their institutional knowledge to variant annotation. Different regions of the genome may require different querying strategies. As users develop these querying strategies, Knome facilitates the exchange of query panels between users, allowing expertise to be shared.
The company says that it is recruiting customers and other experts to develop panels that will run on the knoSYS. These panels can then be tailored by recipient labs to reflect their own preferences and adjusted to accommodate differences that arise from variation in production processes.
Knome’s software runs on a high-performance computing grid, installed directly at the customer site. “We specifically did not pursue the cloud computing option,” says Dr. Salisbury. “Many institutions are wary of putting their sequence data in the cloud. On-site computing also avoids wasting valuable time transferring enormous raw data files to remote servers.”
The hardware component of the knoSYS is expandable, designed to grow as the lab’s testing volume expands.
“Another critical feature of our system, and any system that will be successful going forward,” said Dr. Salisbury, “is the ability to re-query stored genomic sequence. That includes capturing information updates that alter interpretation of earlier analyses, and the ability to run new panels, new tests, as new concerns arise for an individual.”
Currently, the company is focusing on Mendelian diseases and cancer-related somatic variations. Knome has designed the knoSYS as a modular system that can incorporate new databases and align and call solutions. One module that has already been incorporated is the align and call software developed by business partner Real Time Genomics, which uses clever algorithms to accelerate analysis and incorporates joint calling and pedigree information to increase variant calling accuracy.
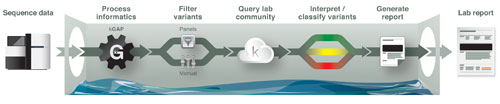
Genome interpretation systems assist labs in absorbing reads and producing reports. One such system, the knoSYS™100 by Knome, integrates an interpretation application and informatics engine with a high-performance grid computer. The diagram shows the system’s workflow.
Pharmacogenomic Analysis
“Is whole-genome/exome sequencing ready for clinical pharmacogenomics applications?” asks Elaine Lyon, Ph.D, medical director of molecular genetics at ARUP Laboratories. “Whole-genome sequencing generates results that we are not yet able to interpret in the context of drug efficacy or adverse reactions. A combination of variants in a single gene or across multiple genes may alter the pharmacogenomic response, or it may be affected by genome-independent factors such as age, other medications, clinical status, and diet.”
ARUP offers pharmacogenomics testing (PGx) as a part of an extensive list of tests. “PGx analysis generated from whole-exome/genome sequencing has a number of challenges,” continues Dr. Lyon. The response is not observed until the individual is challenged with the drug, which doesn’t lend itself to analysis pipelines to filter and prioritize variants for rare diseases. Some PGX genes have pseudogenes, structural variants (gene conversions), and copy-number variants that are difficult to analyze by next-generation technologies.
Instead of whole-exome sequencing, ARUP scientists focus on individual genes and variants that are known to affect drug metabolism. In addition to tests for drug-metabolizing enzymes, ARUP offers single-gene tests based on opioid receptors, oncoproteins, and host factors affecting treatment for infectious diseases. ARUP is also evaluating larger PGX gene panels by next-generation sequencing.
Sequencing data is carefully analyzed and annotated using pharmacogenomics databases. “Databases of clinically relevant mutations are continuously updated,” notes Dr. Lyon. “We plan to contribute to and use ClinVar, an NIH-based public archive that combines relationships among human variations and phenotypes, with supporting evidence for or against pathogenicity.”
ARUP has also developed software to help in prioritizing variants likely to cause particular phenotypes. The program creates a “functional damage score” by assessing whether a variant damages the gene that contains it, and whether the variant is in a gene relevant to the phenotype.
The software will support the ARUP’s offering of Exome Sequencing with Symptom Guided Analysis. The testing aims to provide the etiology of a medical condition if it is suspected to be genetic in origin. Exome sequencing results are prioritized based on the physiological symptoms. Other incidental findings are also analyzed to identify potential pathogenic mutations leading to associated syndromes consistent with recommendations from the American College of Medical Genetics.
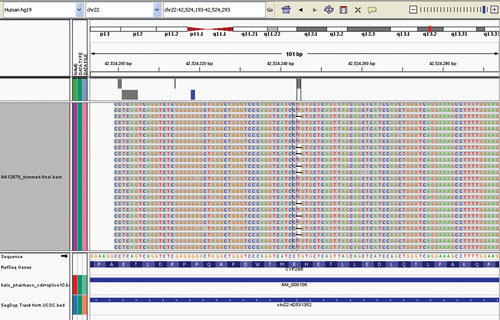
ARUP Laboratories offers pharmacogenomics testing as part of an extensive list of tests. This display, produced by ARUP using the Integrative Genome Viewer, reveals a single base deletion in about half of the analyzed sequence reads, showing heterozygosity for CYP2D6 *3 (c.775delA).