January 1, 2010 (Vol. 30, No. 1)
Bruce Carlson Publisher Kalorama Information
Systems Poised to Break Out of Research Labs into Pharma Industry and Clinical Settings
There has long been interest in sequencing DNA, but until a few years ago there was not much to say about the competitive dynamics of the market for equipment used to perform that sequencing. Mid-decade, the market declined somewhat as most of the potential customers had already invested in high-end sequencers. That changed in 2006 when new technology in the form of second-generation systems reinvigorated competition.
Driving near-term sales of sequencers now is the arrival of third-generation systems, a multiplication of sequenced genomes, and government funding from the stimulus bill. In the long-term, as yet untapped clinical markets also presage a bright future for these systems.
The scientific usefulness of DNA sequencing continues to be proven, and the number of sequenced and catalogued genomes has grown more than five times from where it was at the middle of the decade. Some of these sequencing projects currently under way are quite ambitious and aim to sequence thousands of individuals, tumors, or other unique samples.
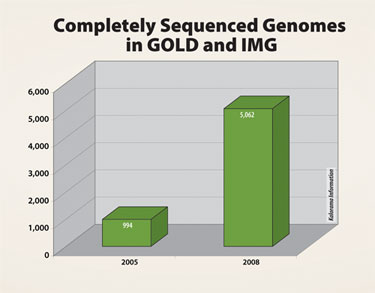
Completely sequenced genomes in Genomes On-Line Database (GOLD) and Integrated Microbial Genomes (IMG)
Increased NIH Funding
The annual budgets for sequencing projects at the NIH and DOE have risen at unprecedented rates in the last few years. NIH is still the key funding agency and funds more than one-third of the projects in the Genomes On-Line Database. This year, new funds have been added as a result of the stimulus bill. In addition to the funding for sequencing projects, the National Human Genome Research Institute has been providing millions of dollars per year to a large number of groups for their development of “$100,000 Genome” and “$1,000 Genome” sequencers.
All of this funding has generated significant momentum in the industry. The last two years have seen fairly significant technology improvements—paired-end techniques, multiplexing, and increasing read-lengths. These advances have resulted in a growth of applications for DNA sequencers and also increased sequencers’ output per hour.
Most of the second-generation sequencers have read-lengths in the range of 25 bases to 75 bases. This would be a severe limitation if the companies were not offering paired-end reads, which allow the fragments to be mapped more easily. All of the sequencing companies are now offering paired-end reads. These have been developed with long inserts of thousands of kilobases. Companies have also been increasing the systems’ read-lengths and adding multiplexing capabilities.
New techniques have definitely amplified the usefulness of second-generation systems. Applications are now shifting away from whole-genome studies and toward more targeted resequencing experiments with fewer genes in larger numbers of samples.
Market Projections
In order to determine a market projection, several factors were considered including the nature of government funding, technology and technology fatigue, and competitive forces. The market is still in a highly volatile state, and new developments are coming along constantly, making it an unpredictable time for all of the companies involved in the market.
The pace of innovation could also prove to be a market limiter. The introduction of second-generation systems has enabled new levels of productivity and new experiments, causing rapid adoption of the products. But there is a flip side to the constant innovation—the demand could slow if labs experience growing pains with the new sequencers.
Many are still trying to address bottlenecks such as the data-management and data-analysis issues. The situation is aggravated because competition has increased, and the end-users are faced with a variety of evolving dilemmas and trade-offs. One technology usually isn’t sufficient, but using multiple sequencer technologies creates integration and workflow issues.
The depreciation of instruments, data management, labor, and other factors has caused the cost of sequencing to be higher than the often-quoted figures, which are related mostly to reagent use. After sorting through some unforeseen issues with second-generation systems, customers are looking out for pitfalls before starting this process all over again with third-generation sequencers.
The high price, performance, and variety of second-generation sequencers could result in a broad first-to-market effect over the third-generation systems; most likely, some labs will simply postpone purchases but will eventually move to the newest technologies. While the new systems have their own performance improvements, they require labs to set up, change, and/or fine-tune their procedures, re-train their personnel, and keep tabs on the different products.
Given these considerations, Kalorama Information believes that the market for sequencers of $480 million in 2008 will peak at $640 million in 2012, then roll downward a bit to $600 by 2014.
We do not believe that sequencers will be limited to research applications forever. The pharmaceutical industry is expected to become a strong customer segment in the near future, due to lower cost and the realization that personalized medicine, biomarkers, and other related post-genomic approaches are likely to help shrinking pipelines.
In the longer term, given the way the science is evolving, the largest opportunity for sequencing technologies may actually be in the doctor’s office or hospital. Some envision portable sequencers within the next 10 years, which does not seem completely unreasonable.
In the near term, it is more likely that existing products will be adapted to specific clinical applications, which has already occurred, for example with HIV drug resistance testing or HLA typing. In order to move the new technologies beyond this early stage, companies will need to be aware of the regulatory processes and other pitfalls of the market. This is ideally addressed through partnering with established companies in the diagnostics industry.
Bruce Carlson ([email protected]) is publisher of Kalorama. Web: www.kaloramainformation.com.



