January 1, 2012 (Vol. 32, No. 1)
Deepak Dibya, Ph.D.
Jeremy Kenseth, Ph.D.
Pierre Varineau, Ph.D.
New Automated CE System Aims to Improve Access to High-Quality Sequence Data
Current next-generation sequencing (NGS) platforms use a variety of technologies including pyrosequencing, ion-sequencing, sequencing by synthesis, or sequencing by ligation. With minor variations, all these systems share a similar DNA library preparation procedure, which includes genomic DNA quality and quantity assessment, DNA fragmentation followed by fragment sizing, and platform-specific adaptor ligation.
Bottlenecks in DNA library preparation are a major problem for NGS labs. A recent survey of 120 labs by Kalorama Information found that library preparation was the main bottleneck in the NGS sequencing process.
One labor-intensive step in DNA library preparation is the size determination and quantification of both un-sheared genomic DNA and downstream fragmented DNA. Existing methods for DNA fragment analysis include agarose gel electrophoresis, chip-based electrophoresis, and capillary electrophoresis.
Agarose gel electrophoresis is labor intensive, requiring gel preparation, sample transfer via pipetting, and image analysis. The images often give distorted or unreliable data, and a separate, second method (UV or fluorescence spectroscopy) is required for quantification.
Chip-based electrophoresis provides faster run times and improved data quality compared to agarose gel electrophoresis, but requires hands-on processing for cleaning, priming, and the loading of gel, markers, and samples onto the system. The samples are analyzed sequentially in the same separation channel, without flushing or priming between injections, which can result in trapped particulates or air bubbles that adversely affect subsequent separations.
Capillary electrophoresis (CE) offers advantages over both agarose and microchip electrophoresis in that gel fill and sample loading are automated. However, many commercial CE instruments are prohibitively expensive and focus on single-strand rather than double-strand DNA analysis. Other CE instruments lack the sensitivity, dynamic range, and separation quality required for adequate DNA library analysis. Although all techniques can be interfaced to a robotic system, only the most sophisticated core labs can afford both the cost and expertise required to operate such systems.
Advanced Analytical Technologies recently introduced the Fragment Analyzer™ CE System. With the capability of analyzing 12 or 96 samples per run, the Fragment Analyzer bridges the gap between a low-volume stand-alone instrument and one that is interfaced with a robotic system. The instrument features:
- an automated capillary flush with fresh gel between every injection cycle to prevent sample carryover;
- the ability to run up to three 96-well sample plates unattended, with a capacity of 1,500 samples/day;
- the ability to switch between two different gel types automatically between injections, enabling multiple sizing ranges to be assessed from one run to the next;
- high separation resolution; and
- high sensitivity detection with a dynamic range for DNA smears of <50 pg/µL to >5,000 pg/µL.
The Fragment Analyzer can be utilized for both quantification and qualification of intact genomic DNA, fragmented DNA, ligated DNA, and enriched DNA. Figure 1 shows an overview of typical NGS library-preparation stages.
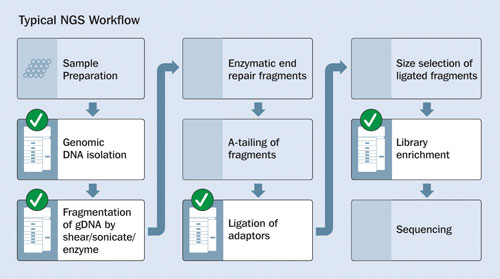
Figure 1. Stages of NGS library construction: Checkmarks indicate where quality/quantity analyses may be performed by the Fragment Analyzer Automated CE System.
Genomic DNA
Genomic DNA (gDNA) is extracted, nonfragmented DNA from cellular matter. Although the gDNA is intentionally fragmented in the downstream construction of NGS DNA libraries, this process relies on high-quality incoming gDNA with a known concentration.
Traditionally, two separate methods are required to fully characterize gDNA:
1) The quality of genomic DNA is monitored using agarose gel electrophoresis, with good quality nonfragmented gDNA defined as a single band at a size of approximately 20,000 base pairs (bp) or higher. Poor quality gDNA is observed as a general broad smear below 20,000 bp.
2) The quantity of genomic DNA is measured using UV-Visible or fluorescence spectroscopy.
The Fragment Analyzer measures both the quality and quantity of intact gDNA with a single automated measurement. Figure 2 shows the analysis of three different samples of gDNA present as intact, partially degraded, or fully degraded forms. The quantity of gDNA obtained with the Fragment Analyzer is equivalent to results obtained with standard fluorometric methods.
In Figure 2, the highly degraded gDNA sample is observed as an extensively shifted smear below 1,000 bp in size. While the fluorometric result indicates a similar concentration, the quality cannot be determined via fluorometry without the use of a secondary technique such as agarose gel, which requires additional cost and time to complete.
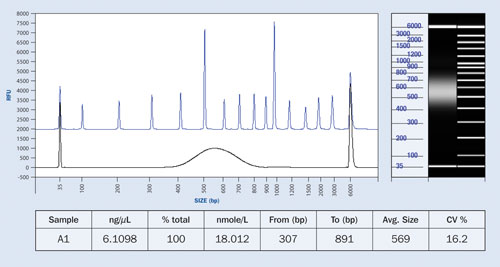
Figure 3. NGS library preparation compared to separation ladder depicting the size distribution of the fragments: The table shows the predicted concentration of the sample.
NGS Fragment Size Determination
All NGS methods employ a technique called shotgun sequencing, whereby the DNA of an organism is randomly sheared into small fragments that are sequenced and combined using a sophisticated computer assembly program. DNA library preparation for NGS employs a controlled fragmentation of gDNA into smaller fragments with average sizes generally ranging from 50 bp to 1,500 bp.
In order to obtain high-quality sequencing data, the size of the sheared DNA must be within a critical range, which is dependent on the NGS platform and method employed. Thus, qualification of the fragments is important. Likewise, many laboratories require the quantity of DNA fragments to fall within a critical concentration range for their process, so quantification of the fragments becomes vital.
The Fragment Analyzer system is able to assess both the quantity and size distribution of NGS fragment libraries across a wide sizing range in a single measurement. The dynamic range is from <50 pg/µL up to 5 ng/µL, with more concentrated samples analyzed via appropriate predilution. The sizing range is from 50 bp to 5,000 bp. Figure 3 shows the concentration and size distribution of a typical NGS library sample.
Advanced Analytical’s Fragment Analyzer technology enables significantly reduced labor for NGS DNA library qualification through method consolidation and automation, with the ability to handle up to 1,500 samples/day.
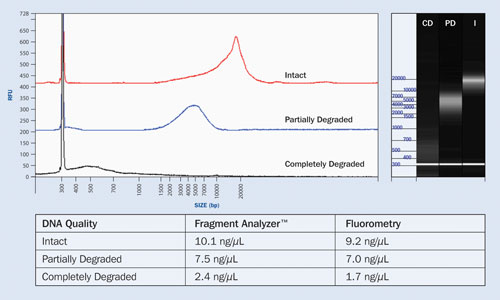
Figure 2. Quality and quantity analysis of intact, partially degraded, and completely degraded genomic DNA samples
Deepak Dibya, Ph.D. ([email protected]), is principal scientist, Jeremy Kenseth, Ph.D., is director, consumer products, and Pierre Varineau, Ph.D., is CTO at Advanced Analytical Technologies.







