April 1, 2009 (Vol. 29, No. 7)
Vicki Glaser Writer GEN
Technology Set to Compete with Microarrays in This Application Arena
Microarray-based gene-expression profiling technology hardly seems like it has been around long enough for a new approach to come along and challenge its market dominance. Yet, there is a growing sense that digital gene-expression profiling, a fully quantitative approach for gene-expression analysis, will replace microarrays in this application area.
Within this emerging market sector for digital RNA counting, the methodologies and technologies are evolving so rapidly that traditional sequencing-based serial analysis of gene-expression approaches are being challenged by more direct RNASeq techniques, driven in large part by the advances in and declining cost of next-generation sequencing technologies.
Digital gene-expression (DGE) technologies are emerging that eliminate the need for restriction enzyme digestion of DNA samples, PCR-based genomic amplification, and ligation of sequence tags. These innovative strategies are not only making whole transcriptome analysis feasible and cost-efficient, they are also creating new commercial opportunities, including the analysis of newly discovered populations of small RNAs believed to have an important role in gene regulation, protein expression, and cell function.
DGE offers distinct advantages over array-based gene-expression analysis systems for transcriptomics, including better coverage, the ability to measure low-abundance genes and to find unknown transcripts, and minimal background noise for increased sensitivity.
A group of Dutch researchers compared deep sequencing-based gene-expression analysis using the Illumina whole genome sequencer to five microarray-based platforms. They concluded “that deep sequencing provides a major advance in robustness, comparability, and richness of expression profiling data.” The authors predicted that, “with the continuously increasing number of reads at reduced costs, RNASeq will become affordable for standard differential gene-expression analysis.”

Inside the Illumina Genome Analyzer, an optical lens is shown scanning a flow cell.
Transcriptome Analysis
Compared to microarray-based methods, the paramount advantage of DGE is that the transcripts are counted and “you do not depend on semi-quantitative analysis of light signaling intensity,” says Björn Rotter, head of functional genomics at GenXPro. “You also don’t need to normalize the data to the same extent, and there is no possibility of false positives, resulting in more reliable information,” he adds.
How DGE is defined depends on who you are talking to. DGE does not describe a technology per se, rather, it is a strategy. In essence, it refers to methods that generate a digital count of gene expression, enabling quantitative differential gene-expression analysis. Initially, DGE was used to describe the SAGE approach, which involves end tagging of cDNA fragments to build an inventory of short sequence tags that can then be sequenced and counted. But the definition of the term has expanded to include methodologies that do not involve end tagging of restriction enzyme digests or require PCR amplification.
The patented SuperSAGE digital gene-expression platform and SuperTAG technology developed by GenXPro rely on a specific 26-base pair tag for accurate transcriptome analysis. SAGE technology can identify what gene is transcribed, how many times a gene is transcribed, and what transcript isoforms are present by counting the number of sequence-specific tags for each individual mRNA.
“The longer the tag, the more reliable the annotation,” says Rotter. GenXPro supplements its intellectual property on SuperSAGE with a patented mechanism to avoid PCR-introduced bias. SuperSAGE is designed to work with Illumina’s Genome Analyzer system and the Roche/454 Genome Sequencer instrument, and GenXPro is experimenting with the Applied Biosystems’ SOLiD™ system.
The main advantage of tag-based gene-expression profiling technology is its open-architecture platform, allowing researchers to detect every transcript present in a sample, including new unexpected transcripts, or antisense transcripts. “It is possible to have a look at rare transcripts, which would get lost in the background noise on microarrays,” Rotter says. In fact, he adds, most of the transcripts in a cell “are rare—present in only one to five copies.”
Applied Biosystems (ABI), a division of Life Technologies, offers both SAGE and RNASeq kits on its SOLiD system next-generation sequencing platform. The company’s SAGE product incorporates a 27-base pair tag and has a dynamic range of 5 to 6 logs—a 1,000-fold improvement over the typical 2.5 to 3 log readouts achieved with microarrays, according to Roland Wicki, director of SOLiD strategy. That translates to greater sensitivity and the ability to see more targets with SAGE sequencing technology compared to microarray-based methods.
However, states Wicki, “RNASeq, or a whole transcriptome shotgun RNA sequencing methodology, is the more compelling approach,” and he anticipates that in the future it will likely supplant both microarrays and SAGE methods. The power of whole transcriptome sequencing is defined by what it can enable, including expression analysis of specific exons and detection of splice variants, alternative transcripts, and novel splice junctions. RNASeq can also be used to analyze allele-specific expression and measure the expression levels of heterozygous SNPs. The same strategy and data can yield information on the full spectrum of mutations in a sample and can be used to detect and quantify fusion transcripts.
“None of this can be done on microarrays— it is all new,” and requires whole transcriptome analysis capability, adds Wicki.
In a presentation at the Association of Biomolecular Resource Facilities meeting, a team of scientists from ABI and from the University of Cambridge (U.K.), described a method of “Deep Sequencing-Based Whole Transcriptome Analysis of a Single Cell” using the ABI SOLiD System. The researchers combined next-gen sequencing with whole transcriptome amplification to develop a digital gene-expression profiling assay at single-cell resolution.
They demonstrated that this single-cell cDNA deep-sequencing assay was able to detect expression of thousands more genes than a cDNA microarray technique. Furthermore, for genes detected by both strategies, the sequencing approach yielded novel transcript variants for many of the genes, suggesting that the transcriptome of a single cell is more complex than previously realized.
In February, ABI began delivering the new SOLiD 3 system to customers. It provides simplified workflows and greater accuracy, according to Wicki. Using two-base encoding—the system reads every base twice, providing a means of internal error correction—it can differentiate between real mutations and misreads by the instrument. The new model also has higher throughput and can analyze 400 million tags, or sequence reads, of 50-mer fragments in a single run. R&D runs have demonstrated the potential of the instrument to process up to one billion tags/run.
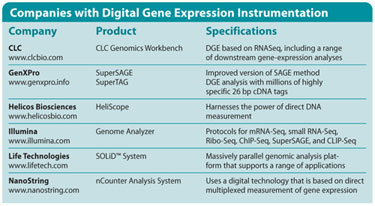
Companies with Digital Gene Expression Instrumentation
Single Molecule Sequencing
Patrice Milos, Ph.D., vp and CSO at Helicos Biosciences, described the company’s genetic analysis system at Cambridge Healthtech’s “Next Generation Sequencing” conference in San Diego last month in a presentation entitled, “Enabling True Biology with Helicos™ Single Molecule Sequencing.” DNA sample preparation on the HeliScope™ Single Molecule Sequencer requires no ligation steps or PCR amplification, Dr. Milos said.
The basic process for quantitative RNA analysis involves creating a single-stranded copy of the total or poly A+ RNA sample using reverse transcriptase, adding a polyA tail to the cDNA, and allowing these to hybridize to a flow cell surface containing oligo dT to initiate sequencing by synthesis reactions of the single-copy transcript tags. The result is both sequence information and read counts for the transcripts present in the sample.
By eliminating the need for PCR, the Helicos technology offers “a truly unbiased view of biology,” said Dr. Milos. As an example she described transcriptome analysis of a human placental sample: “You will get well over 14,000 genes reported that range from very rare—1 to 2 transcripts/million—up to 100,000 transcripts/million. The 50-channel HeliScope can generate 12 to 14 million reads in a single channel, with a resolution in the range of one transcript/million.”
Dr. Milos reported that customers are beginning to use the Helicos system for a broad range of applications that combine accurate quantitation and sequence information, such as copy number variation (CNV) analysis, gene-expression profiling of different tumor types or cell lines, and discovery of novel transcripts.
Whereas, the current HeliScope can accommodate in the range of 12 to 14 million sequence tags per channel, Dr. Milos expects to see a rapid increase in throughput by about 2½-fold in the near future. Average read lengths, now 34–35 bases, will also increase.
“We are now enabling paired reads on the Heliscope,” she said. “This improvement allows the instrument to read two regions of a molecule in a single run, with the same sample preparation and will make it possible for customers to do more exploration at the whole genome level.”
A team of Swedish researchers from Uppsala University recently presented a random array format and associated decoding scheme for “targeted multiplex digital molecular analyses” in Nucleic Acid Research. This method analyzes DNA samples using sets of padlock or selector probes that identify target sequences and create circular DNA molecules, which are then amplified via rolling circle amplification.
Padlock probes are “linear oligonucleotides that become circularized in a strictly target-dependent ligation reaction.” Selector probes are similar to padlock probes, with “target-specific ends for target recognition, flanking a DNA sequence with elements for amplification,” but they differ in that they “are designed to hybridize to the end-sequences of restriction digested genomic DNA fragments and thus template DNA ligase assisted circularization of specific genomic DNA sequences.”
Sequence Assembly Solutions
DNAStar recently introduced QSeq, the first product to use the company’s disk sort alignment (DSA) algorithm for quantitative RNASeq applications and digital gene-expression experiments.
“I believe that RNASeq is in the validation stage now,” says Tom Schwei, vp, GM, and CFO of DNAStar. “As we prove that the sequence-based techniques are at least as good as, if not better than the microarray-based techniques, there will be more uptake of RNASeq and digital gene expression. I think that component of microarrays [gene-expression analysis] will go down proportionally as the RNASeq methods gain acceptance.”
DNAStar also introduced a beta version of SeqMan NGen (SNG), which can support next-generation sequence assembly on multiple instruments, including those of Illumina, Roche/454 Life Sciences, and Helicos, as well as Sanger sequencing data. The SNG algorithm assembles genomic fragment data up to 150 megabases in size and integrates with Lasergene sequence analysis software for sequence analysis and visualization.
SNG can assemble an entire human chromosome, according to Schwei, and the largest project the company has tackled so far is human chromosome 8, assembling 150 megabases (of Roche/454 sequence data) with 7x coverage on a desktop computer in less than three hours.
“We believe the software will be able to handle 30–50 megabase genomes on a template basis with 10 to 20 times coverage,” Schwei says. He emphasizes that the algorithm can be used for either template or de novo type sequencing projects.
Clusters of interest within Heat Maps generated with DNAStar’s ArrayStar software can be selected and stored as gene sets for additional evaluation.
Improving Sensitivity
The nCounter® Analysis System from NanoString Technologies uses a digital technology based on molecular barcodes and single molecule imaging to achieve direct multiplexed measurements of gene expression. The system includes a fully automated sample-prep station, a digital analyzer, CodeSet barcodes, and the necessary reagents and consumables.
nCounter technology creates a series of unique tags, which differ by the combination and position of four colors. Using four colors and eight possible tag positions, for example, 48 or 65,536 unique tags can be created. Each tag is chemically linked to a target-specific probe (a sequence-specific oligonucleotide) to produce a reporter probe. The reporter probes are then pooled to form a CodeSet that is mixed with a sample, such as a total RNA extract, and allowed to hybridize in a single reaction.
Removal of unhybridized probes and subsequent imaging of the bound individual reporter probes yields a count of the number of copies of each code, and thus of each gene, that is present in the sample. In comparative studies, the company showed the nCounter system to be more sensitive than microarray-based methods and similar in sensitivity to real-time PCR.
Earlier this year the FDA put out a call for volunteers to participate in the Sequencing Quality Control (SEQC) project, intended “to objectively assess the technical performance of different next-generation sequencing technologies in DNA and RNA analyses and to evaluate the advantages and limitations of various bioinformatics solutions in handling and analyzing the massive new data sets.”
The FDA views SEQC as a natural extension of its MicroArray Quality Control project, which evaluated and compared various microarray-based methods and technologies for gene-expression profiling.







