October 1, 2010 (Vol. 30, No. 17)
Shannon Morey Kinney, Ph.D.
Jurate Bitinaite, Ph.D.
Sriharsa Pradhan, Ph.D.
Approach Strives to Make Hydroxymethylated and Methylated Cytosine Quantification Reproducible
Epigenetics refers to the study of heritable changes in genome function that occur without a change in the primary DNA sequence of an organism. One such change is DNA methylation, most frequently characterized by an enzymatic modification at the fifth position of cytosine (5-mC), present abundantly in the context of CpG dinucleotides. The formation and maintenance of 5-mC is catalyzed by cellular DNA (cytosine-5) methyltransferases (DNMTs).
The biology of 5-mC and its role in epigenetic inheritance and gene expression are well known. In addition to 5-mC, mammalian DNA also contains a variety of other modified nucleobases at low levels, which arise by DNA damage, normal metabolism, and other environmental factors. These modifications are generally removed during the onset of a new cell cycle.
Recent observations involving mouse Purkinje and granule neuron cells identified an additional modified cytosine, 5-hydroxymethylcytosine (5-hmC). This modified base was also found in the genome of undifferentiated embryonic stem (ES) cells. A family of genes encoding Tet (ten eleven translocation) proteins (Tet1, Tet2, and Tet3) catalyze the oxidation reaction of 5-mC to 5-hmC.
In addition to this mechanism, the transfer of formaldehyde to cytosine by DNMTs may be another cause of 5-hmC formation. The biological role of 5-hmC is the subject of much current speculation. A popular hypothesis is that 5-hmC may be an intermediate between 5-mC and cytosine in the mammalian genome. For example, conversion of 5-mC to 5-hmC and subsequent deamination of 5-hmC to 5-hydroxymethyluracil (5-hmU) would generate a mismatch between two opposite bases (5-hmU:G), resulting in activation of mismatch repair pathways.
In another study, a reversible enzymatic reaction catalyzed by DNA (cytosine-5) methyltransferases led to the release of formaldehyde from 5-hmC, producing unmodified cytosine. Therefore, through this mechanism, 5-hmC may participate directly in DNA demethylation.
Furthermore, disruption of the Tet1 and Tet2 genetic loci has been reported to associate with hematologic malignancies. A fusion of human Tet1 with the histone methyltransferase MLL has been identified in several cases of acute myeloid leukemia (AML) associated with the t(10;11)(q22;q23) translocation.
Homozygous null mutations and chromosomal deletions of the Tet2 locus have been discovered in myeloproliferative disorders, suggesting that Tet2 may function as a tumor-suppressor protein.
The cytosine nucleobase can reside as either unmethylated or methylated, including hydroxymethylated (5-mC and 5-hmC) forms. Methylated cytosine is generally associated with gene repression.
Several biomarkers rely on methylation status as a disease prognostic and/or diagnostic marker (e.g., methylation of Sept9 as a biomarker for colon cancer). However, neither gene expression nor biomarker studies have taken the presence of 5-hmC into account as, currently, there are no routinely used technologies that can distinguish 5-mC from 5-hmC.
The gold-standard approach to detect methylated cytosine is bisulphite conversion followed by DNA sequencing. Although this chemical method can reliably identify cytosine versus 5-methylcytosine, it does not distinguish between 5-mC and 5-hmC.
EipMark 5-hmC and 5-mC Analysis Kit
To further the study of 5-mC and 5-hmC in DNA, New England Biolabs (NEB) has developed a robust, simple method that utilizes beta-glucosyltransferase (beta-GT), an enzyme that glucosylates 5-hmC in the presence of UDP-Glucose (UDP-Glc), and two methylation state sensitive restriction enzymes (MspI and HpaII) that differ in their ability to cut methylated DNAs containing a CpG dinucleotide.
To definitively determine whether a specific locus is 5-hmC modified, DNAs are treated with beta-GT, modifying all 5-hmC to glucosylated 5-hmC (5-ghmC) (Figure, step 1a). A control reaction is also set up without beta-GT, leaving all 5-hmC modifications intact (Figure, step 1b).
Digestion of the two reactions with MspI can be used to differentiate between 5-ghmC and 5-mC modified cytosines, as MspI will cleave 5-mC and 5-hmC, but not 5-ghmC (Figure, steps 2a and 2d). Digestion of the beta-GT treated sample with HpaII further enables the determination of the percentage of total methylation (5-mC, 5-hmC, and 5-ghmC), as all three modifications block HpaII cleavage (Figure, step 2b).
Finally, a control of beta-GT treated uncut DNA should be analyzed to determine the total amount of the DNA of interest (Figure, step 2c). Following digestion, qPCR with primers flanking the DNA sequence of interest can be used to quantify levels of 5-hmC and 5-mC at the given locus (Figure, step 3).
Amplification is dependent on the levels of MspI and HpaII cleavage and, therefore, is a direct correlation of the quantities of 5-hmC and both 5-mC and 5-hmC, respectively. Validation of this method is shown in this article using synthetic DNAs with different ratios of the various states of cytosine methylation.
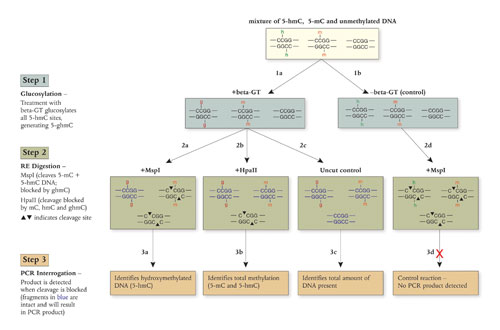
The workflow for detection and quantitation of 5-hmC using the EpiMark 5-hmC and 5-mC Analysis Kit: The DNA of interest is treated with beta-glucosyltransferase (beta-GT) and UDP-Glucose (UDP-Glc). Beta-GT transfers glucose from UDP-Glc onto 5-hydroxymethylcytosine (marked as “g”). MspI cuts DNA containing 5-hydroxymethylcytosine but does not cut if it is glucosylated; in contrast, HpaII is blocked by any of the modifications of cytosine. After PCR amplification, if the CpG site of interest contains 5-hydroxymethylcytosine, a PCR product will be obtained after beta-GT treatment and MspI digestion. A reaction containing DNA that has not been treated with beta-GT and is also digested with MspI serves as a negative control.
Results
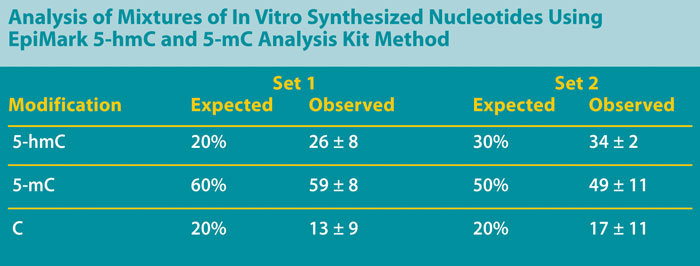
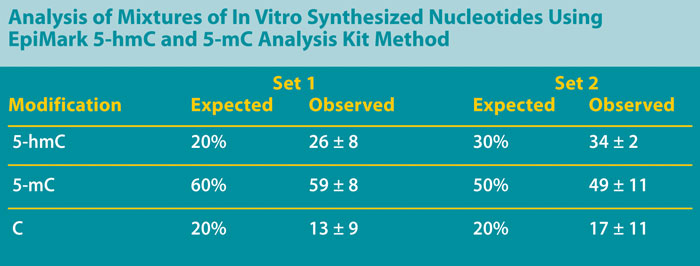
In order to verify that the above method can accurately detect differences between a population of differentially methylated DNAs, mixtures of in vitro synthesized oligonucleotides that are unmethylated (C), methylated (5-mC), or hydroxymethylated (5-hmC) were prepared. Two heterogeneous mixed sets of these sequences were assessed: set #1 contained a mixture of 20% C / 60% 5-mC / 20% 5-hmC, and set #2 contained a mixture of 20% C / 50% 5-mC / 30% 5-hmC.
Both sets were treated with beta-GT followed by digestion with MspI or HpaII. Undigested DNA served as a positive control for amplification, while the digestion of nonglucosylated DNA with MspI served as a negative control. Quantitative PCR of the synthetic DNA yielded results consistent with the input amounts of the differentially methylated DNAs in each sample (Table).
These results indicate that this method reliably differentiates between unmethylated, 5-mC, and 5-hmC modified DNAs, and allows the quantitation of the various levels of each.
Analysis of mixtures of in vitro synthesized nucleotides using EpiMark 5-hmc and 5-mc analysis kit method
Conclusion
Quantification of locus-specific epigenetic modifications in the mammalian genome is an integral part of understanding epigenomic changes during development and disease. The EpiMark 5-hmC and 5-mC Analysis Kit from New England Biolabs offers a simple, reproducible, and robust method to examine the methylation status of a DNA population; addresses the need for detection and quantitation of 5-hmC in genomic DNAs; and opens up opportunities to determine whether 5-hmC modifications of DNA can be exploited in the identification of novel biomarkers.
Further, quantitative PCR can be replaced by end-point PCR, providing scientists with a simple “yes or no” answer to whether a given loci contains 5-hmC. Lastly, multiplexing of this protocol for end-point PCR may be performed for different amplicons using standard thermocyclers.
The EpiMark method followed by qPCR analysis is currently being applied to study the differences in hydroxymethylation patterns for a variety of biologically relevant modifications in mammalian DNAs during the different stages of development. In unpublished studies, researchers at NEB have shown variations in 5-hmC levels of genomic DNAs extracted from brain, heart and liver tissue samples from murine and human.
Shannon Morey Kinney, Ph.D. ([email protected]), is a post-doctoral research associate, Jurate Bitinaite, Ph.D. ([email protected]), is a senior scientist, and Sriharsa Pradhan, Ph.D. ([email protected]), is the division head of RNA biology at New England Biolabs.