November 1, 2015 (Vol. 35, No. 19)
Kathy Liszewski
The Assay Focuses Its Inner Eye on Complex Biology, Through 3D Models, Reporter Genes, Ubiquitin Traps, and More
If the eye is the lamp of the body, the assay is a searchlight that illuminates complex biology. Yet there is a significant difference between the inner light and the assay, which has an inner light that is merely luminescent.
In contemplations of the inner light, singleness of vision is what counts. Assays, however, must shift their gaze as necessary. In fact, an assay in development is like a searchlight sweeping its beam, following and isolating its targets. A well-aimed assay, then, is one that reveals information needed for a particular kind of analysis, an analysis that may, for example, advance the discovery, development, or validation of a drug, or permit the assessment of a therapeutic intervention.
While it may swivel and adjust its focus, the better to scan complex biological landscapes, the assay, no less than a searchlight, must have some stability. Assays must behave predictably with precision and specificity, often in a high-throughput setting. New strategies and methodologies in contemporary assay development include the creation of 3D models that more accurately capture cellular microenvironments; the use of primary cells to accomplish phenotypic modeling; the engineering of responsive, division-arrested cells; and the targeting of specific cellular pathways.
The environment in which a tumor develops plays an important role not only in its metastasis, but also in its response to drugs. “Most drug discovery programs involve high-throughput screening (HTS) using cultured cancer cell monolayers,” informs Hilary A. Kenny, Ph.D., an assistant professor of obstetrics and gynecology at the University of Chicago. “This approach, however, does not faithfully recapitulate the complex inner architecture, cellular heterogeneity, and critical interactions of the tumor’s microenvironment.”
For a case in point of drug discovery lacking sufficient environmental context, consider the underwhelming results that have been achieved against ovarian cancer, an aggressive, often fatal illness in which tumor cells detach from the female genital tract and metastasize via peritoneal fluid to the surface of abdominal organs.
“While new drugs have appeared on the market, the five-year survival rate has not changed for two decades,” Dr. Kenny notes. “We wanted to mimic the structural organization and function of the main site of ovarian cancer metastasis in an assay amenable to HTS. Using the knowledge obtained from many other 3D models, we built an organotypic HTS assay.” In this assay system, Dr. Kenny mimics the surface histology of the omentum and peritoneum by mixing primary human omental cells from healthy donors (mesothelial cells, fibroblasts) with extracellular matrix material.
“The primary cells are isolated, plated, and grown for one week then immediately used in the assay or stored in liquid nitrogen for use months or years later,” details Dr. Kenny. “For assaying, we plate the organotypic culture and grow for 48 hours before adding cultured human ovarian cancer cells bearing a green fluorescent protein tag. After 16 hours, the unattached cells are washed away, and the adhesion/invasion of the ovarian cancer cells is measured by fluorescent spectroscopy.
“HTS is performed in up to 1,536-well plates, and to date more than 48,000 compounds have been screened with z-factors from 0.4 to 0.6 in collaboration with the National Center for Advancing Translational Sciences.”
For the future, Dr. Kenny feels this type of 3D assay could be applied to other peritoneal cavity cancers such as colon, gastric, and pancreatic cancers. She recalls that when she first presented her data in meetings, there was apprehension about running a multicellular assay in a 1,536-well plate. She is optimistic, however, that doubts will fade in light of recent results.
“Excitingly, this is a proof that more complex assays should be approached and developed,” she insists. “This platform can be used to perform consistent, reproducible, and quantitative HTS for identifying effective drugs more quickly, as well as for reducing attrition rates.”
Validating 3D Cellular Assays
Although 3D cell culture can provide a more physiologically relevant environment, it can also add complexity, presenting challenges to experimental design and interpretation. “Most cell-based assays were developed and optimized on 2D monolayers in standard cell culture,” maintains Terry Riss, global strategic marketing manager, cell health, Promega. “Thus, there is an unmet need for assays validated for use with the larger 3D culture systems.”
The validation of 3D culture systems means addressing a number of routine questions. Will the reagents effectively lyse all 3D structures? Will cell lysis destroy the target marker? Will reagents penetrate to the center of the culture? Will the mass of cells block signal detection? Such questions prompted Promega to evaluate the effectiveness of various assay chemistries.
“Based upon our studies and collaborations,” we decided to reformulate a cell viability assay we developed to measure ATP,” explains Dr. Riss. The assay now incorporates a higher detergent concentration, more physical disruption (a shaking period of 5 minutes), and a longer lysis period (30 minutes). We now provide the CellTiter-Glo® 3D Cell Viability Assay.
“We also modified an apoptosis protocol we developed to measure caspase-3/7 activity. The assay now incorporates longer incubation times (30 minutes versus 5 minutes) and more shaking. Generally, we now recommend changing the detergent formulation when possible, increasing physical disruption (that is, shaking), as well as a longer incubation in the lysis solution.”
Another approach is to avoid lysis altogether and instead utilize assays employing small molecules that penetrate larger microtissues. “One example is the use of DNA-binding dyes,” Dr. Riss points out. “We also have applied the RealTime-Glo® MT cell viability assay in 3D systems. For this assay, an added prosubstrate diffuses into cells. Only live cells release a substrate that is used by luciferase to produce light.”
According to Dr. Riss, a take-home message is that when one is using 3D cultures, it is “imperative” to carefully optimize and validate assays with each cell type and each 3D model system.

Validation of assay reagents applied to 3D culture models requires a thorough consideration of complete penetration of reagent through multiple cell layers. Validation of this kind can be accomplished with Promega’s CellTiter-Glo 3D Cell Viability Assay, a homogeneous method to determine the number of viable cells in 3D cell culture based on quantitation of the ATP present.
Phenotypic Assays in Primary Cells
Phenotypic screening is a well-honed and classical approach to drug discovery. Indeed, this approach has led to a disproportionally large number of first-in-class drugs with novel mechanisms of action. To enhance its phenotypic screening capabilities, contract research organization Charles River Laboratories acquired the services division of Galapagos. The purchase included the Galapagos’ BioFocus business unit, which developed PhenoFocus, a portfolio of more than 100 complex human primary cell-based assays that span more than 20 different disease areas.
“We use cells derived from tissue, blood, or differentiated stem cells to provide the most relevant biology for developing high-throughput assays that aid drug discovery,” says Jamil Aarbiou, Ph.D., a team leader at the BioFocus unit that Charles River currently maintains in Leiden.
In addition to the human primary cell-based assays, the company has developed more than 300 drug discovery assays and performed more than 100 screening campaigns comprising more than 30 million test wells. “We also develop and perform client-specific assays,” notes Dr. Aarbiou. “These may include RNAi-based screens, tests of drug candidates on patient-derived cell models, and target-based screens.”
Two of the challenges faced with phenotypic screening and testing are variability among donors and the need for low passage number of patient-derived cells. “To overcome this variability, we assess similar cell models from a number of donors,” explains Dr. Aarbiou. “We also can search for donors that meet client-specific needs in order to provide the best and most reliable disease model.”
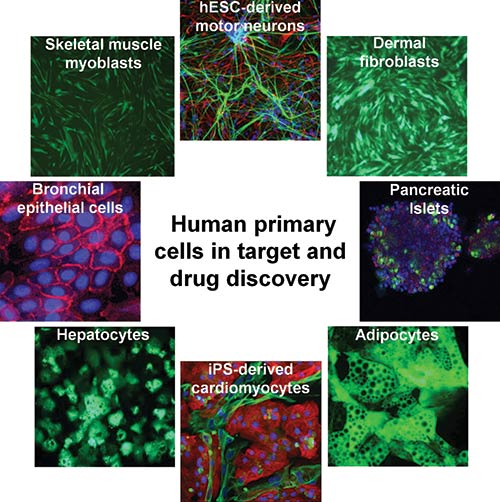
Charles River Laboratories’ BioFocus business unit uses its PhenoFocus portfolio to advance drug discovery. The portfolio comprises assays that use cells derived from tissue, blood, or differentiated stem cells. Through PhenoFocus, over 100 complex human primary cell assays are available for hit finding, target discovery, and target validation.
Reporter Gene Assays
Reporter gene assays have been around for many years. This technology, however, has been updated by Biomonitor for enhanced specificity, sensitivity, and elimination off-target effects. “Cell engineering is a powerful technique for the development of sensitive, precise, and highly specific assays,” says Michael G. Tovey, Ph.D., Biomonitor’s managing director. “We have engineered reporter gene cell lines that can quantify the activity of a range of biopharmaceuticals.”
The company’s reporter gene assay is called iLite. Basically, it embodies the engineering of two reporter constructs into specific cells. The cells express the Photinus pyralis firefly luciferase under the control of a drug-responsive promoter and the Renilla reniformis sea pansy luciferase under the control of a constitutive promoter.
This technology allows both luciferases to be assayed sequentially in the same well of an assay plate. Also, drug-induced luciferase activity can be normalized relative to Renilla luciferase expression. Provided as assay-ready frozen cells, the technology is amenable to HTS, and assays can be run in two to five hours.
“These engineered drug-responsive division-arrested reporter gene cell lines respond predictably under assay conditions,” asserts Dr. Tovey. “Thus, a main advantage for these assays is that these same cell lines can then be used in drug discovery, for quantifying drug potency, and to analyze the functional drug levels and antidrug neutralizing antibodies.”
The iLite technology includes assays for a number of interleukins, interferons, and TNF-α, and it can be used to quantify antibody-dependent cytotoxicity and other attributes.
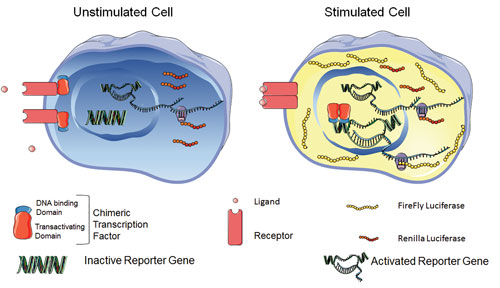
On a cell engineered with Biomonitor’s iLite reporter gene technology, a target interacts with a drug. A signal transduction pathway is activated, as is (in turn) a synthetic chimeric transcription factor, which then binds to a DNA sequence regulating transcription of the firefly luciferase (FL) reporter gene. Drug-induced FL activity is normalized relative to expression of Renilla luciferase under the control of a constitutive promoter, rendering assay results independent of cell number, serum matrix effects, or nonspecific toxicity. [Michael Tovey, Ph.D.]
DUBs and Drug Development
The multifaceted molecule, ubiquitin, is a small (~8.5 kDa) but powerful regulatory protein expressed by most eukaryotic tissues. It can become attached to a protein subtrate via ubiquitination, a kind of post-translational modification. After it undergoes ubiquitination, a protein substrate can show various effects. Addition of a chain of ubiquitins (polyubiquitination, often via the ubiquitin lysines K48 and K29) is the “molecular kiss of death,” as it leads to degradation via proteosomes. Other polyubiquitinations (for example, via K6, K11, or K63) or addition of single ubiquitin proteins may regulate processes such as inflammation, DNA repair, endocytic trafficking, and translation.
Opposing the role of ubiquination is the family of deubiquitinating enzymes (DUBs) that remove and recycle ubiquitin from substrate proteins. DUBs include cysteine proteases and metalloproteases that cleave the amide bond between ubiquitin and the substrate protein. DUBs, however, are also capable of activating ubiquitin. With these wide-ranging activities, many DUBs have been linked to diseases such as cancer by virtue of their oncogene-stabilizing abilities.
“DUBs are increasingly recognized as drug targets for a variety of diseases,” says Daniel Cardillo, application scientist, global discovery applications, PerkinElmer. “DUB assays, however, are difficult to translate into sensitive HTS assays.”
“There are many limitations of current DUB assays,” he continues. “Most employ nonnatural synthetic fluorescent substrates. The many wash steps are time-consuming, and assays often have higher cost, lower data quality, and detect only specific binding partners.
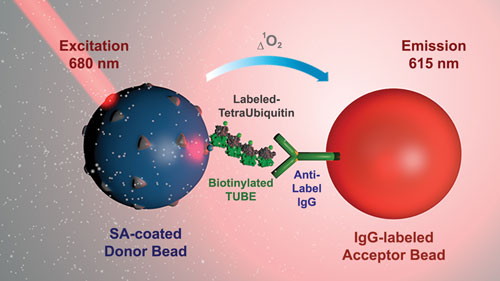
“In an effort to provide screening assays that better recapitulate the natural environment of the target, we developed a novel linkage-specific biochemical DUB method that utilizes Tandem Ubiquitin Binding Entities (TUBEs) coupled to our AlphaLISA platform, a bead-based no-wash antibody technology that uses an amplified luminescent proximity homogeneous assay.”
The TUBEs serve as “ubiquitin traps” that bind to tetraubiquitin substrates of at least four natural linkages. Cardillo asserts that this type of approach is both simpler and more natural: “All reagents are added in one well without the need for washing steps. Larger molecules can be used. There is a high signal to background and Z factor. Finally, using larger complexes allows more robust, biologically relevant assays.”
PerkinElmer plans to release assays for five of the eight natural linkages by the end of the year. Then it will continue developing the remaining three.
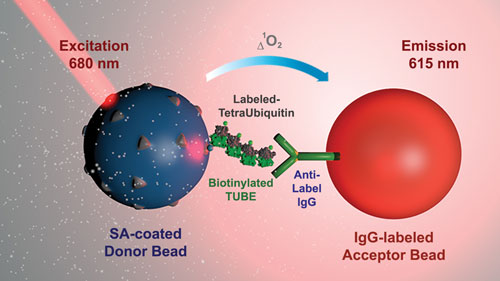
PerkinElmer has enhanced its AlphaLISA platform so that it may serve as a linkage-specific biochemical deubiquitinase (DUB) assay. AlphaLISA, a bead-based no-wash antibody technology that uses an amplified luminescent proximity homogeneous assay, is now equipped with Tandem Ubiquitin Binding Entities (TUBEs), which provide “ubiquitin traps” that bind to tetraubiquitin substrates of at least four natural linkages.
Novel Thermal Stability Assay
Another new assay blends thermal denaturation-based methods with homogeneous biochemical ligand binding assays. “It is well established that ligand binding protects proteins from thermal denaturation; however, adapting this concept to easily measure quantitative ligand potencies has been a challenge,” reports Daniel Treiber, Ph.D., vice president, KINOMEscan division, DiscoveRx.
DiscoveRx’ newly released methodology is designed to provide quantitative readouts of an inhibitor’s half-maximal effective concentration (EC50). “We’ve developed a new technology platform, called HitHunter® Pulse, that expands on existing thermal denaturation-based methods for ligand-binding applications,” explains Dr. Trieber. “This novel approach uses a simple protocol and provides quantitative data for inhibitor potency rank ordering and HTS hit validation.”
HitHunter Pulse capitalizes on two DiscoveRx technologies: enzyme fragment complementation (EFC) and pulse denaturation. In EFC, the catalytic domain of the protein of interest is tagged with an enhanced ProLabel (ePL). The ePL binds to and complements the enzyme acceptor fragment (EA), and the resultant active beta-galactosidase enzyme turns over substrate to produce a chemiluminescent signal.
If the target protein is denatured, however, complementation is impaired, and chemiluminescent signal is lost. Ligand binding protects the protein from denaturation and thus preserves the chemiluminescent signal.
The second technology, pulse denaturation, incorporates a gentle series of brief heating and cooling cycles. Pulse denaturation improve the quantitative power of HitHunter Pulse over standard denaturation methods, which use a single aggressive denaturation step.
According to Dr. Treiber, this new patent-pending platform addresses the problem of how to build more quantitative denaturation-based assays that are simple to execute: “The pulse approach can improve assay sensitivity by ~10-fold over standard denaturation protocols. We have validated the assay with known inhibitors, where we measured accurate potency rank orders. We are preparing to release a number of kits for targets such as bromodomains, kinases, and methyltransferases.”
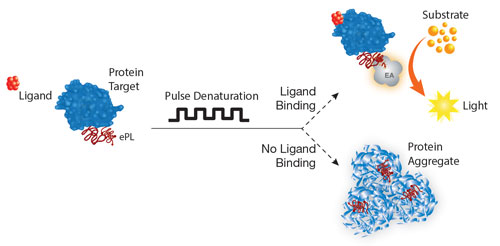
DiscoveRx’ HitHunter® Pulse biochemical compound binding assays can quantify inhibitor potencies and support hit validation and lead optimization activities during high-throughput screening through its enzyme fragment complementation and pulse denaturation technologies.
Establishing Immunogenicity and Pharmacokinetic Bioequivalence
During biosimilar drug development, biotech companies often confront this conundrum: “Is it better to use two separate immunogenicity (or pharmacokinetic) assays to analyze the reference product and the biosimilar, or should one assay be used?”
The latest FDA guidance document on biosimilarity issued in April 2015, “Scientific Considerations in Demonstrating Biosimilarity to a Reference Product (Section D2—Clinical Immunogenicity Assessment),” outlines a single-assay bioanalytical approach. The one-assay strategy provides the most comprehensive and reproducible information in the most cost-effective way while minimizing the inherent variability associated with using two separate assays, noted the agency.
To assess immunogenicity similarity, AIT Bioscience recommends employing one competitive inhibition analysis using biosimilar and innovator therapeutics on the same plate (in separate wells) in parallel during the assessment of the confirmatory cut point in method validation and Tier 2 Confirmatory testing of sample analysis. To provide a more reliable assessment of potential anti-drug antibodies, AIT Bioscience typically establishes the cut point using matrix from both healthy patients and patients with disease.
Though the latest FDA guidance offers no recommendation for pharmacokinetic (PK) assays, AIT Bioscience suggests using a one-assay approach for PK analysis for the same scientific and cost considerations described previously. It’s the most reliable and straightforward path to demonstrate comparable analytical similarity as recommended by Marini et al.
Another key message from the April 2015 document is the importance of preclinical animal studies. AIT Bioscience recommends conducting an animal study that compares both PK and immunogenicity responses to provide an assessment of biosimilarity prior to initiating a costly and lengthy clinical study. Minor structural differences can dramatically affect a biotherapeutics’ efficacy and/or safety.
A clear difference in preclinical immunogenicity could foreshadow an unanticipated structural difference between the biosimilar and innovator therapeutic. Thus, animal studies can reveal vital information about the biosimilarity of a biotherapeutic to help guide future clinical studies.