September 1, 2007 (Vol. 27, No. 15)
Paul Siebert Ph.D.
Generating Sufficient Amounts of Representative cDNA for Analysis
Most clinical tissue samples are preserved using formalin fixation followed by embedding in paraffin. By some estimates, almost 90% of tissue sections are prepared using this methodology. Although this process is excellent for maintaining morphological features, it does not preserve RNA well. In fact, RNA integrity in formalin-fixed, paraffin-embedded (FFPE) tissue is poor. As a result, it is difficult to extract sufficient intact RNA for analysis.
There is much interest in analyzing gene expression in these clinically important samples. For these reasons, there is a strong need to have a reliable and robust approach to extract and prepare for expression analysis the small amount of intact RNA from FFPE-prepared tissues.
Although there are ways to extract RNA from FFPE samples, the degraded character of this RNA makes it difficult to reliably amplify it to produce enough usable material for expression analysis. Most amplification procedures, such as T7 in vitro transcription (T7 IVT), rely on poly-A priming and produce product that is highly biased toward the 3´ end of transcripts. These amplification methods do not typically amplify anything beyond the last 600 to 1,000 bases of the transcript. In addition to leaving much of the RNA inaccessible to analysis, these 3´-biased approaches are particularly problematic with the degraded RNA derived from FFPE tissues, where they typically cannot produce usable product.
mRNA Amplification from FFPE Tissues
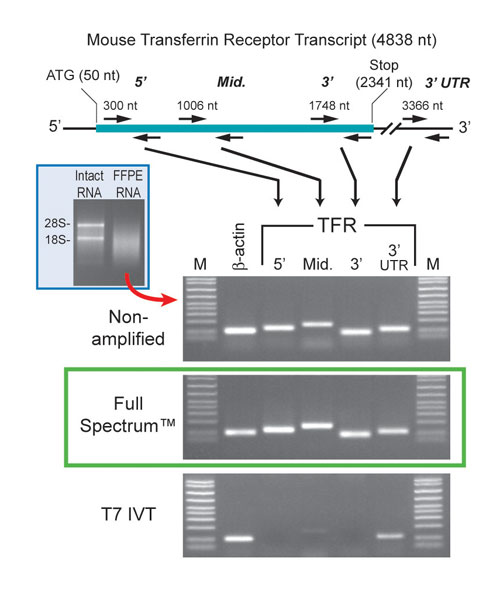
To test the ability of System Biosciences (www.systembio.com) Full Spectrum™ Kit to amplify RNA derived from FFPE tissues, the company isolated RNA from two 20-µm sections of mouse liver. The RNA from this tissue is substantially degraded (Figure 1). Following amplification, we ran samples of the cDNA product on a gel. The results show a range of cDNA products that are all relatively small in size.
The company also amplified a similar amount of RNA using T7 IVT. As anticipated, the amplified products look similar. However, since the Full Spectrum method does not rely on poly A priming like the T7 IVT approach does, the Universal Primer should amplify all regions of the transcripts relatively equally.
Thus, 5´ portions of the transcripts should be present in the Full Spectrum product. To test this, System Biosciences amplified both samples with the transferrin receptor primers whose locations are shown in top part of Figure 1, along with b-actin control primers. The results support the conclusion that all regions of the transcripts are amplified relatively equally using the Full Spectrum RNA Amplification Kit (Figure 1, bottom).
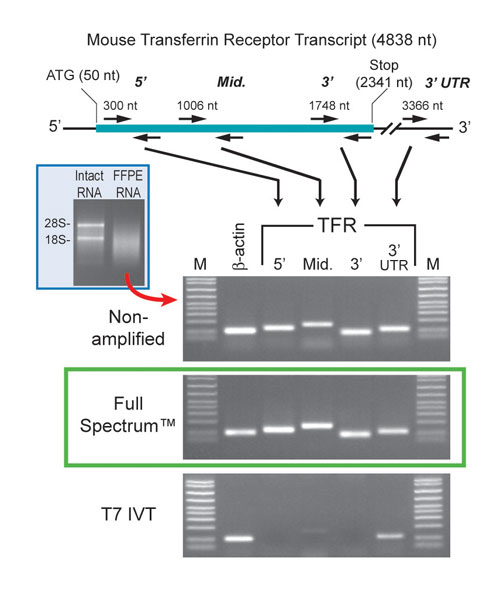
Figure 1
Relative Transcript Quantities
Any RNA amplification method must amplify all transcripts equally to ensure relevant biological expression levels are being measured. To demonstrate the uniformity of amplification achieved with the Full Spectrum RNA Amplification Kit, expression levels of 50 human genes were measured before and after amplification of 20-ng total RNA, and the Ct values were plotted in Figure 2A. The identical experiment was performed using a T7 IVT method with the exception that 50-ng total RNA was used, and the results were plotted (Figure 2B). As shown, the Full Spectrum technique (R = 0.940) produces results that meet and exceed those of the T7 IVT method (R = 0.770). The data supports the assertion that the Full Spectrum technique maintains relative transcript quantities during the amplification process.

Figure 2
Conclusions
These results demonstrate that Full Spectrum Complete Transcriptome RNA Amplification provides an alternative approach to amplify RNA derived from FFPE tissues for expression analysis. In addition to robustly and reliably amplifying this notoriously difficult RNA from these valuable samples in a way that maintains the relative levels of each transcript, use of the Full Spectrum approach ensures that all regions of the transcripts are present in the amplified products. While producing a more representative population, this unbiased amplification also means that Full Spectrum samples can be used for analysis of gene families and alternate transcripts of the same gene. This feature also makes Full Spectrum amplification suitable for the preparation of samples for real-time qRT-PCR and microarray analysis.
Paul Siebert, Ph.D., is director of gene amplification at System Biosciences. Web: www.systembio.com.
Phone: (650) 968-2200
E-mail: [email protected]