July 1, 2011 (Vol. 31, No. 13)
Diane M. Retallack
Russell Coleman
Using a Combinatorial Approach in a P. fluorescens System to Boost Production
Recombinant protein expression is a time-consuming task because it requires the combination of a large number of variables that can often subvert timely development. Highly developed bacterial expression platforms are particularly suited to overcoming many of these obstacles because of the rapid growth rates of these organisms and the large number of expression tools that have been developed to meet expression challenges.
However, because primary amino acid sequences of large proteins reveal little information on how best to express a given protein, deciding which tools or combinations of tools will be effective to produce a particular target protein is often elusive. In particular, the expression of secreted proteins with multiple disulfide bonds can be challenging in a microbial system.
What has been needed is a way to apply parallel processing to expression strain development so that multiple expression strategies can be accessed and tested simultaneously. Pfenex has created a comprehensive Pseudomonas fluorescens-based protein production platform consisting of a broad spectrum of effective expression tools that can be seamlessly accessed and integrated to yield an expression strain producing large amounts of functional protein.
Replacing the traditional, linear, and iterative approach to strain development with a high-throughput, parallel-processing model enables construction and evaluation of 1,000 unique expression strains combining novel gene-expression strategies and host cell phenotypes in about one month’s time.
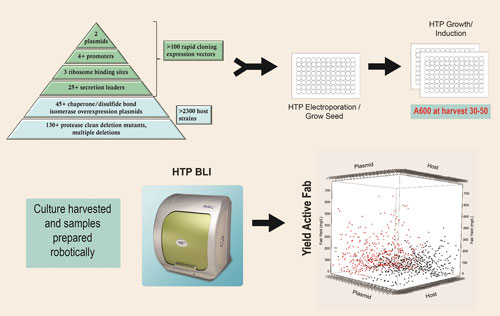
The toolbox pyramid diagram contained in Figure 1 describes the expression strategies and engineered hosts available through the platform that are used to generate thousands of completely unique expression strains. These tools can be seamlessly accessed so that any expression scenario can be combined with any engineered host by 96-well electroporation. Both the expression plasmids combining the expression strategies listed in Figure 1 and the competent host strains are off-the-shelf items.
The expression plasmids themselves all employ the same cloning strategy, utilizing optimized synthetic gene fragments to streamline target gene cloning. It is difficult to predict exactly which combination of secretion leader and ribosome binding site (RBS) will work best for the heavy chain (HC) and light chain (LC) of various Fabs.
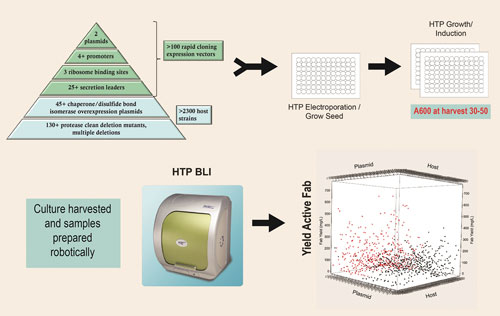
Figure 1. Strain screening process: 1,000 or more unique expression strains are constructed and screened to identify combinations that result in high levels of active Fab expression.
In fact, we have observed that with each Fab expressed, the optimal strain selected for each Fab is composed of different expression strategy/host strain combinations. A number of monocistronic and bicistronic constructs is screened for any given Fab, which is facilitated by the rapid cloning vectors illustrated in Figure 2.
Synthetic fragments designed with HC and LC coding regions, as illustrated in the figure, are cloned into any number of rapid cloning vectors, which are then electroporated into an array of host strains. These hosts may have one or more protease genes deleted and/or may overexpress folding modulators (chaperones, disulfide isomerases), which enable production of high levels of intact, assembled, functional Fab.
The ability to perform all aspects of strain engineering in an automated, high-throughput, 96-well format is key to this combinatorial approach. An obligate aerobe, P. fluorescens, is uniquely suited to growth by simply shaking in a 96-well plate (0.5 mL culture volume) without oxygen supplementation, reaching cell densities of 30–50 A600 units, as described in Figure 1.
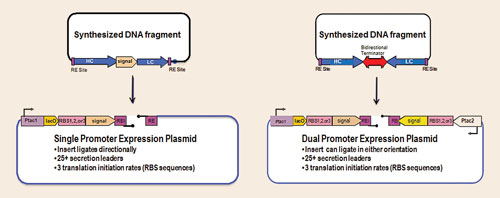
Figure 2. Rapid cloning vectors: monocistronic or bicistronic expression vector libraries employ a single cloning strategy for streamlined fusion of target-encoding genes with a variety of ribosome binding site strengths and periplasmic secretion leader coding sequences.
Bioanalytical Assays
This makes available a significant amount of expressed protein with which to perform bioanalytical assays even at this small scale. Typically, we perform a two-tiered assay scheme. Sample preparation is automated, with harvest, sonication, and separation of soluble and insoluble fractions performed on a single robotic platform.
The first round of analysis is designed to provide information on the titer of active Fab expressed in the soluble fraction, using high-throughput, automated biolayer interferometry. Binding of assembled, active Fab to its antigen can be used to rank the expression strains, rapidly identifying the top yielding 1-5% of the expression strains evaluated. Alternately, binding to protein L or protein A can be used to quantitate soluble Fab.
The scatter plot depicted in Figure 1 is an example of results from the first-tier analysis of a typical 1,000-strain screen. Both host strain and expression plasmid can influence the overall yield of active Fab.
In this example, strains carrying monocistronic expression plasmids are shown in black and those carrying bicistronic expression plasmids are shown in red. The top yielding strains typically produce 100 to 500 mg/L of active Fab at the 96-well scale.
The second tier of assays is performed on selected samples and can include affinity enrichment of active Fab, SDS-CGE, Western blot, and even LC-MS to further assess the quality of the expressed Fab. The ability to perform intact mass analyses at this scale can give us information on the fidelity of signal peptidase cleavage of the secretion leaders on both the heavy and light chains.
All of these steps, from receipt of optimized gene fragments through completion of the analyses and selection of strains to evaluate in fermentation, take approximately five weeks to complete. In the case that the goal is the isolation of research amounts of protein, sufficient amounts of protein can be isolated from shake flask cultures due to the large amount of biomass (i.e., 30–50 A600 units) that can be recovered from such cultures.
When larger quantities of material and/or an optimized production strain is the end goal, up to 10 high-yielding strains, representing a diversity of expression strategies and host strain combinations are selected for fermentation “range-finding” experiments to identify the most robust strain for further development.
Fermentation Conditions
A design-of-experiments (DoE) approach is taken to first evaluate up to nine sets of experimental fermentation conditions at the 10 mL (4 mL working volume) microbioreactor scale. Variables such as temperature, pH, and inducer conditions are studied to identify robust strains and a range of effective conditions for high-level expression. Based on these results, two to three strains are studied in more conventional 1 L reactors to confirm the effectiveness of the conditions identified at the 4 mL scale.
These two rounds of fermentation studies together are accomplished in about four weeks, and at this point in the screening process, strains and conditions producing over 1 g/L of active Fab have often been identified. The fermentation broth available from the 1 L reactors can be used to purify protein for further studies.
Employing a combinatorial approach to strain engineering, we have shown that in little more than two months, 1,000 unique expression strains can be constructed, evaluated, a fermentation process partially developed, and an expression strain selected for further downstream development. This platform has delivered high-quality material for preclinical evaluation more rapidly than any other system, allowing for faster go/no-go decisions to be reached and thus minimizing opportunity costs associated with development.
In several cases, expression of the same Fab in other systems such as yeast or E. coli failed to produce soluble, active Fab. An example is shown in Figure 3, where in about 10 weeks from receipt of the synthetic genes, this parallel screening approach enabled the production of 0.5 grams of >99% pure active Fab for preclinical studies, thus avoiding stalled development that would have occurred had Fab production been attempted in a less robust platform.
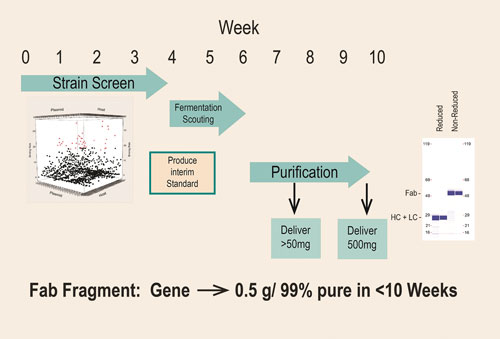
Figure 3. Rapid transition from discovery to development for the production of Fab: In approximately 10 weeks from receipt of a synthetic gene fragment, significant amounts of preclinical material is provided.
Diane Retallack ([email protected]) is director, molecular biology, and Russell Coleman ([email protected]) is senior scientist, molecular biology at Pfenex.